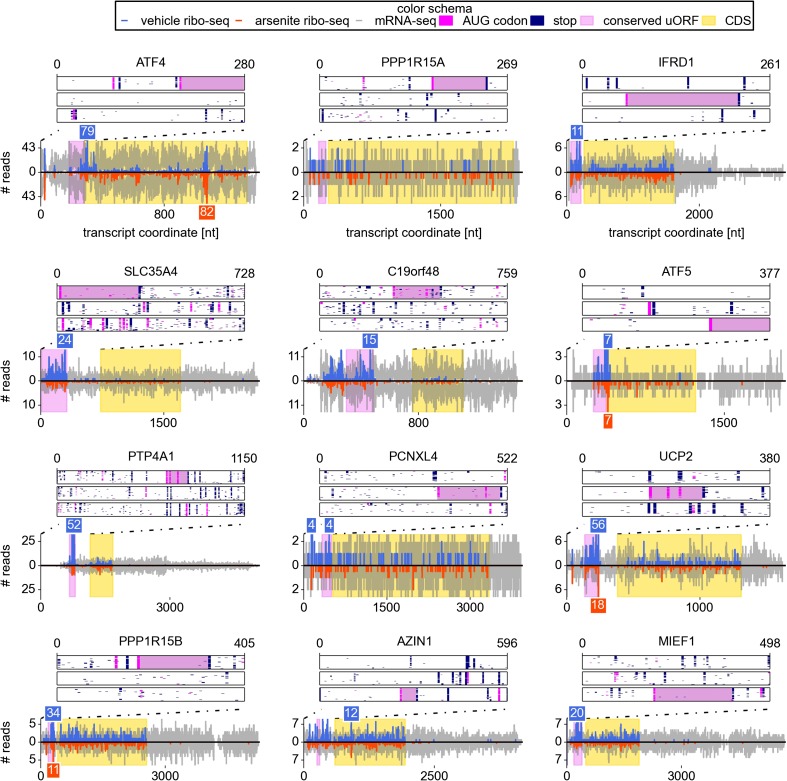
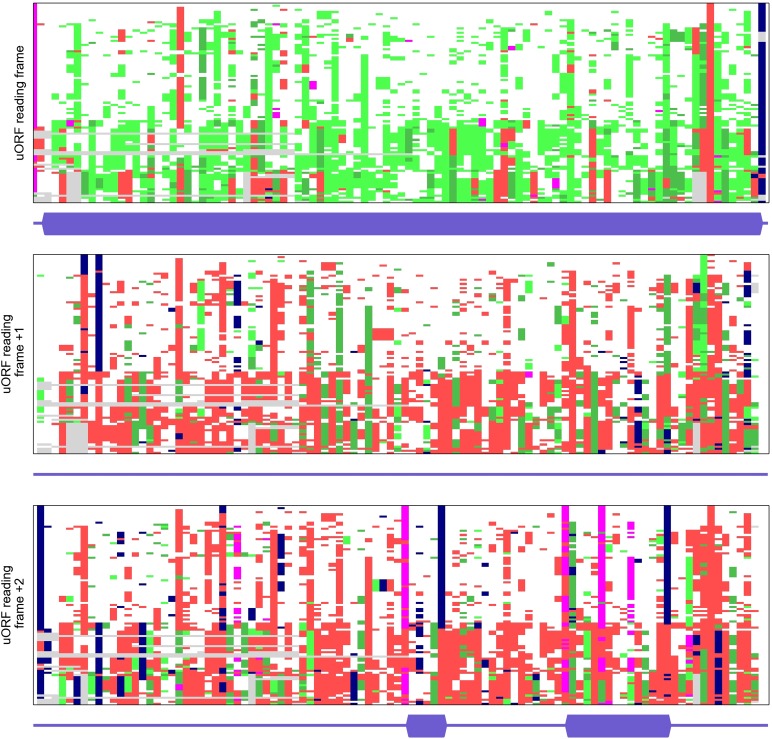
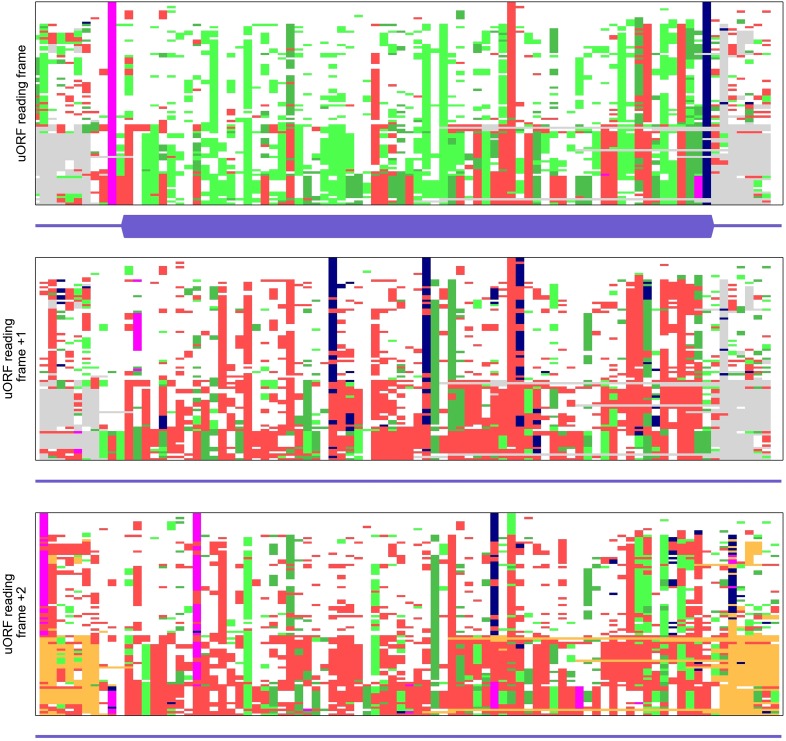
Figure 2. Upstream open reading frame (uORF) conservation and ribosome density for the eight top most stress resistant mRNAs in terms of their translation efficiency and also for mRNAs of UCP2, PPP1R15B, AZIN1, and MIEF1.
Bottom plots for each mRNA show counts of mRNA-seq reads (grey) and ribosome reads (blue and red) as columns (control: positive values; arsenite treatment: negative values). The annotated CDS region is highlighted in yellow. Translated conserved ORFs in the 5′ leaders are highlighted in violet. Read counts above the cut-off are shown with numbers above corresponding columns. Top plots represent conservation of uORF features within the leaders of the orthologous mRNAs (upstream of annotated CDS) obtained from the analysis of genomic alignments of the 46 vertebrates using the human sequence as a reference. Each box corresponds to one of the three reading frames where AUG codons are shown as pink dots and stop codons as navy dots in each of the genomic sequences used in the alignments. Regions of multiple sequence alignment corresponding to translated conserved uORFs are highlighted in violet. Introns and gaps were removed from the alignments.