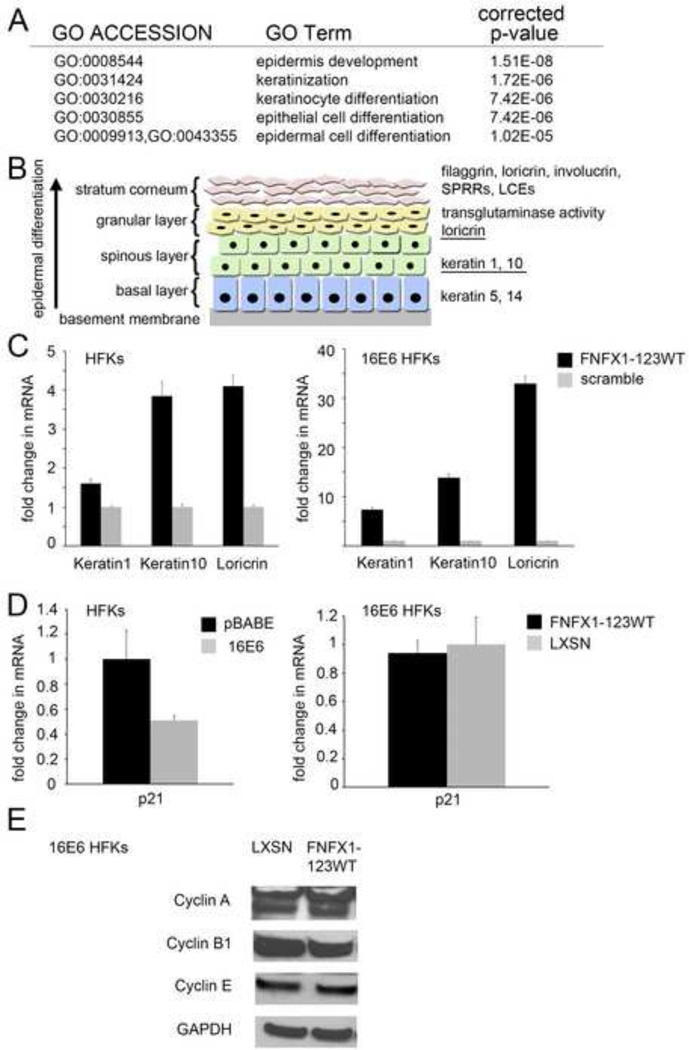
Figure 4. FNFX1-123WT induced differentiation markers in 16E6 HFKs.
(A) Top statistically significant Gene Ontology (GO) terms for Biological Process classification of the differentially increased gene expression comparing 16E6 HFKs transduced with overexpressed FLAG-tagged NFX1-123 wild type (FNFX1-123WT) and isogenic scramble short hairpin RNA control (scramble). (B) Markers of differentiation in stratified squamous epithelium. The expression of keratins and other structural proteins shift with differentiation. (Illustration adapted from (Hoffjan & Stemmler, 2007)). (C) HFKs and 16E6 HFKs transduced with FNFX1-123WT or a scramble shRNA control were analyzed by qPCR. Relative levels of Keratin1, Keratin10 and Loricrin mRNA were calculated using the ΔΔCT method, normalizing mRNA levels to GAPDH within each sample. Values shown are the mean fold change in each sample compared to the scramble vector control. Error bars represent the standard deviation for triplicate samples. (D) HFKs transduced with 16E6 or a vector control (pBABE), and then FNFX1-123WT or a second vector control (LXSN), were analyzed for p21Waf1/CIP1 mRNA levels using the housekeeping gene 36B4 as control. Values shown were the mean fold change in each sample compared to the scramble vector control. Error bars represent 95% confidence intervals from the technical triplicate replicates shown. (E) Representative immunoblot of Cyclin A, B1, and E protein in whole cell extracts of 16E6/LXSN and 16E6/FNFX1-123WT HFKs. GAPDH shown as a loading control.