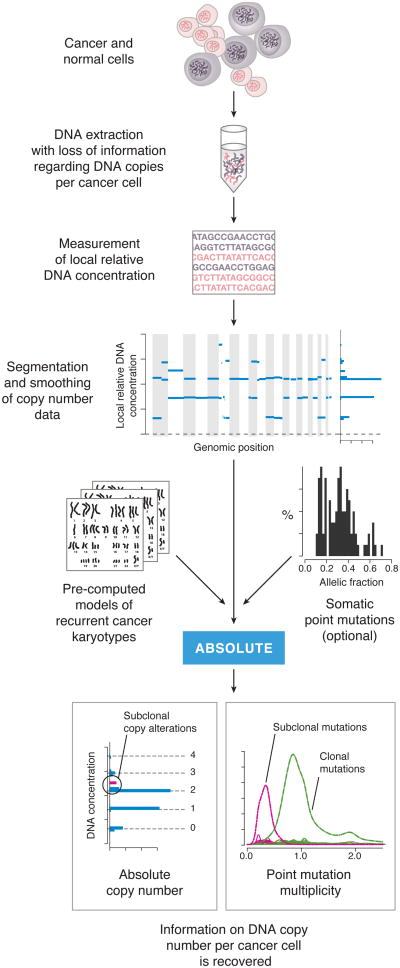
Figure 1. Overview of tumor DNA analysis using ABSOLUTE.
A constant mass of DNA is extracted from a heterogeneous cell population consisting of cancer and normal cells. This DNA is profiled using either microarray or massively parallel sequencing technology, giving a genome-wide profile of DNA concentrations (blue lines). ABSOLUTE uses statistical models of recurrent cancer karyotypes to interpret the DNA concentrations as discrete copy states corresponding to predominantly clonal somatic copy number alterations, although some subclonal alterations are often present. If somatic point mutation data are available (from sequencing of the DNA), then the allelic fractions (fraction of sequencing reads bearing the non-reference allele) of these mutations may be used help to interpret the DNA concentrations. In addition, the allelic fractions may be reinterpreted as integer allelic copies per cancer cell (multiplicity), potentially revealing subclonal point mutations.