Figure 3. Pan-cancer application of ABSOLUTE.
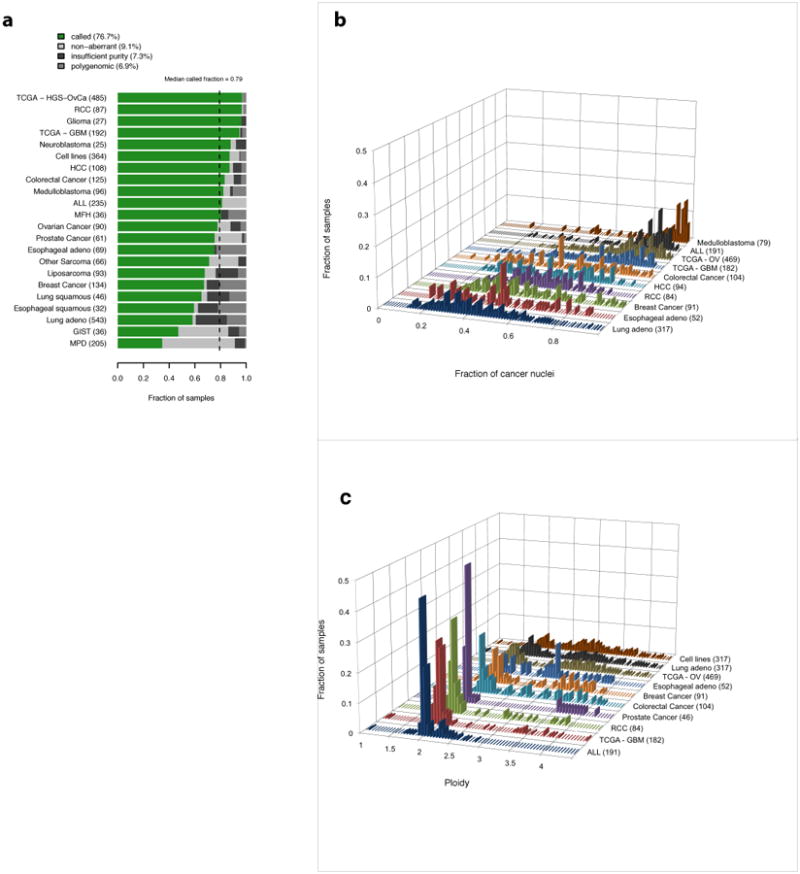
a, ABSOLUTE result types: (i) ‘called’ -- unique purity/ploidy solution; (ii) ‘non-aberrant’ --sample has no detectable somatic copy-number alterations; (iii) ‘insufficient purity’ – insufficient fraction of cancer cells; (iv) ‘polygenomic’ discrete copy-ratio levels could not be determined. See Online Methods and Supplementary Fig. 5 for a description and examples of each result type.
b, Distribution of estimated tumor purity for several datasets. The number of called tumor samples in each group is shown in parentheses. We note that, because heavily contaminated tumors are difficult to call using ABSOLUTE, several of these distributions are biased towards higher purity samples.
c, The number of called tumor samples in each group is shown in parentheses. Because tumors without SCNAs cannot be called using ABSOLUTE, these distributions do not incorporate the prevalence of such samples.
