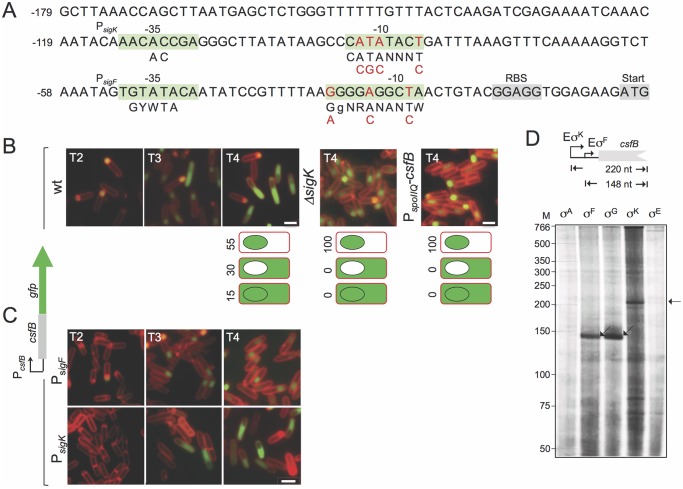
Fig 2. Expression of csfB in sporulating cells.
A: the panel represents the regulatory region of the csfB gene, with the -35 and -10 elements of putative σF- or σK-dependent promoters. The consensus sequences for promoter recognition by σFor σKare represented below the promoter sequence (R is A or G; W is A or T; Y is C or T; N, is any base) [8,9]. The point mutations introduced in the putative σF- or σK-dependent promoters are indicated in red. The ribosome-binding site (RBS) and start codons are also represented, for reference. B and C: the localization of a CsfB-GFP fusion protein, expressed from the amyE locus in a csfB mutant background was monitored by fluorescence microscopy at the indicated times (in hours) after the onset of sporulation in re-suspension medium, in cultures of the following strains: an otherwise wild type, a ΔsigK mutant, a strain expressing CsfB-GFP from the forespore-specific PspoIIQ promoter (in B); strains bearing the point mutations indicated in A, in the σF (PsigK) or σK promoters (PsigF) (in C). Cells were stained with the membrane dye FM4–64 prior to fluorescence microscopy. Scale bars, 1 μm. The cartoons in B represent the percentage of cells with GFP fluorescence in the coloured compartment at hour 4 of sporulation in re-suspension medium. D: in vitro transcription reactions using RNA polymerase containing the indicated B. subtilis sigma factors, and a PCR template containing the csfB promoter. Products of about 148 or 220 nucleotides were expected for transcripts initiating from the putative σF- and σK-dependent promoters, as represented.