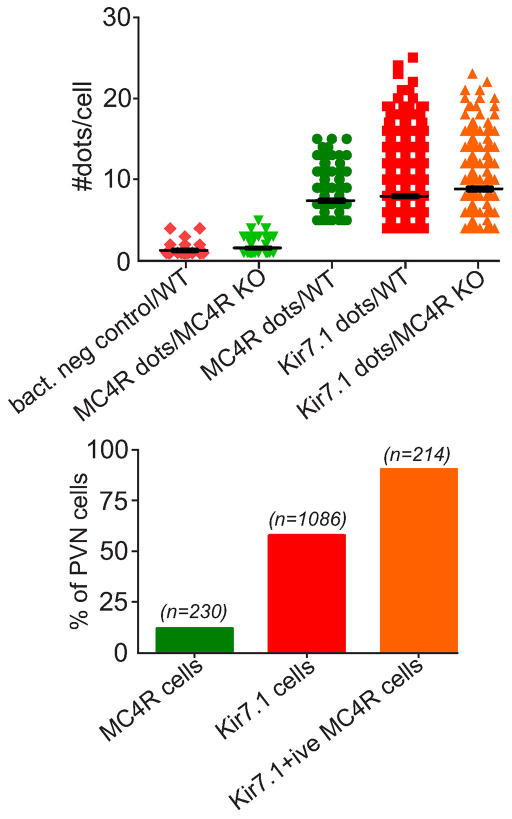
Fig. ED4. Quantitation of MC4R and Kir7.1 RNA in PVN cells.
Single molecule RNA detection in sections was quantitated by counting fluorescent dots associated with individual cells (Fig. ED3). Background threshold was determined from the number of dots per cell in sections resulting from hybridization using a negative bacterial DNA control, or from hybridization of the MC4R probe to sections from the MC4R knockout mouse (a, columns 1 and 2). Threshold-subtracted dot numbers were then used to determine the % of PVN cells expressing MC4R or Kir7.1, and the % of MC4R cells expressing Kir7.1; cells were considered positive if the number of dots exceeded the mean of the negative controls by 3x std (b).