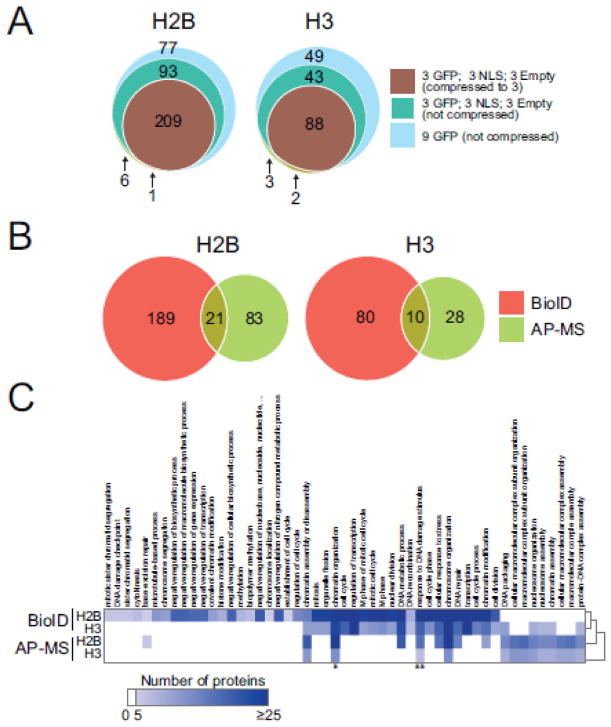
Figure 3.
BioID is successful at mapping interactomes of histones H2B and H3. (A) Venn diagram of significant interaction partners detected by BioID analysis of histone H2B and H3 after running SAINTexpress using different types of background controls and compression parameters. (B) Venn diagram of significant interaction partners detected by BioID and AP-MS (data generated in [3]) analyses of histones H2B and H3. (C) Functional annotation of the significant interaction partners for H2B and H3; the protein count for biological processes (GO FAT) with Benjamini-Hochberg FDR correction of 0.01 or smaller is mapped. Proteins contributing to GO term GO:0051276 (*, chromatin organization) and GO:0006974 (**, response to DNA damage stimulus) are shown in supplementary Figure 2.