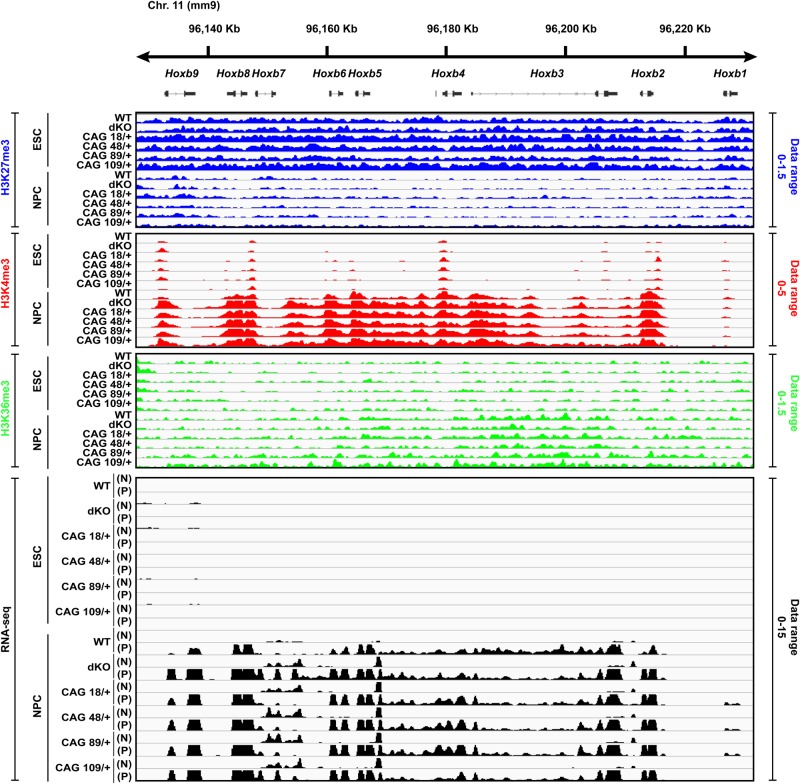
Figure 3.
Hoxb cluster illustrating genome-wide ChIP-seq and RNA-seq analyses. A snapshot of the IGV (http://broadinstitute.org) genome browser view at the location of the developmentally regulated Hoxb cluster (mouse chromosome 11qD) shows the ChIP-seq library-size normalized reads density (see Materials and Methods) for histone H3K27me3, histone H3K4me3 and histone H3K36me3 across the Htt wild-type, Htt null and the four Htt CAG knock-in ESC lines and for the NPC derived from them. Also shown are the RNA-seq reads density with the + strand (P) and – strand (N) indicated. Library-size normalized reads density data range for each histone modification and RNA-seq datasets are indicated on the right side of the tracks. For all six genotypes, after neural induction, the level of histone H3K27me3 at the gene TSSs is decreased with increased enrichment of histone H3K4me3 that is concomitant with RNA expression and enrichment of histone H3K36me3 across the gene bodies, thereby indicating comparable pluripotency and neural differentiation status for the members of the isogenic panel. Additional QC results for the ChIP-seq and RNA-seq datasets are provided in Supplementary Material, Figures S1, S2A and B.