Figure 2.
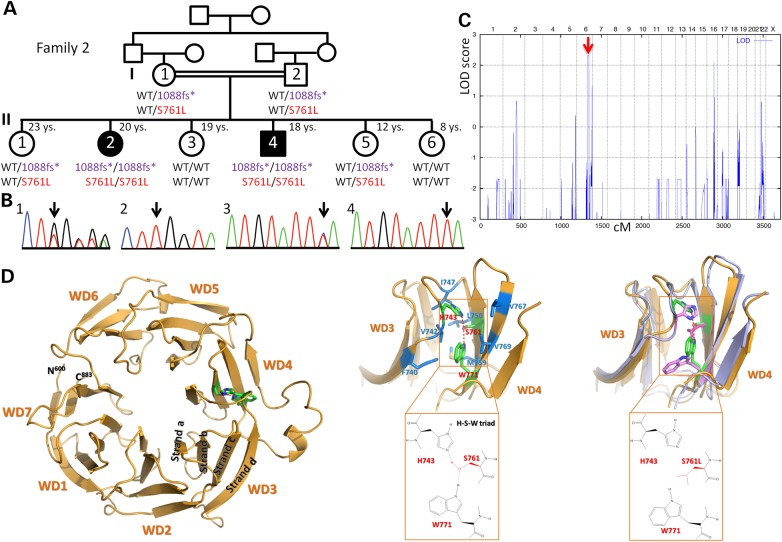
Family 2 with JBTS and two homozygous AHI1 variants. (A) Pedigree (black symbols: JBTS). The sample of II:2 was subjected to WES. A deceased sister and a healthy brother (whose samples were not available for genotyping) are not shown due to space constraints. For clarity, abbreviated codes (1088fs*, S761L) were used for the AHI1 variants p.Trp1088Leufs*16 and p.Ser761Leu. Both homozygous variants co-segregate with the JBTS phenotype. S761L is considered to be the disease-causing mutation and therefore depicted in red. The 1088fs* variant is considered non-pathogenic and therefore depicted in purple. (B) Electropherograms depicting heterozygosity (1, I:1) and homozygosity (2, II:2) for 1088fs*, as well as heterozygosity (3, I:1) and homozygosity (4, II:2) for S761L. (C) Genome-wide homozygosity mapping implicating the parents and all siblings shown in (A) identified a single HBD region with a combined maximum parametric LOD score of 2.31 in both affected siblings, for a region on chromosome 6q23.2–q24.3 that contains AHI1. (D) Left: Structure of AHI1 protein as predicted by the WDSP algorithm. Middle: More detailed structure of the WD3- and WD4-repeats (with some loops deleted for clarity) that show the hydrogen bond network formed by residues H743-S761-W771 (red/green) and its hydrophobic environment (corresponding residues in blue). Right: The superposition of simulated WD3 and WD4 structures for S761 (wild type, green) and S761L (mutant, pink) shows the overall conformational changes caused by the S761L mutation.