Figure 2.
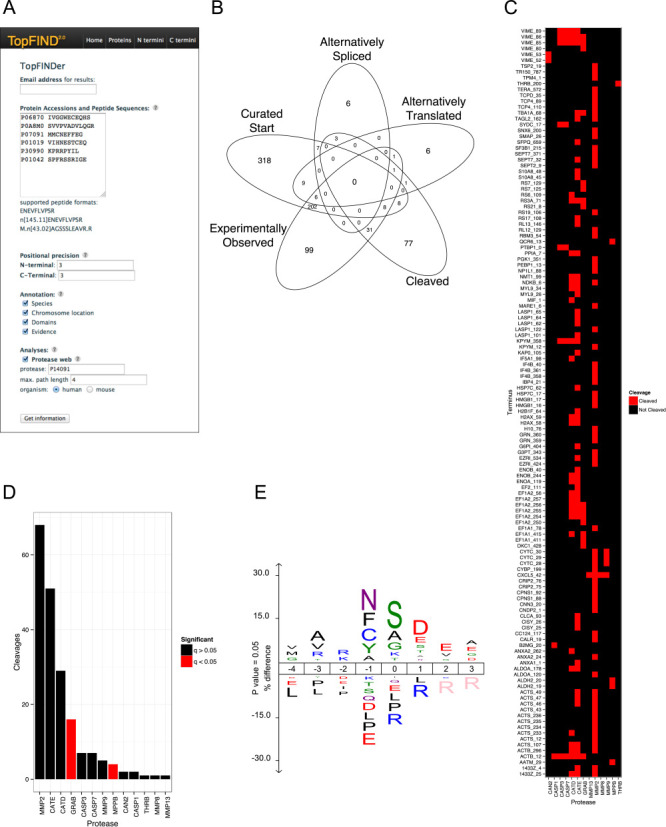
Input and output of TopFINDer. (A) Input mask within the TopFIND web interface. (B) Venn diagram showing the overlap of termini evidences retrieved from TopFIND for a list of proteins. Evidence is either UniProt annotated terminus (Curated Start), terminus of an isoform derived from alternative splicing (Alternatively Spliced), or from alternative translation (Alternatively Translated), from cleavage (Cleaved), or a terminus observed in a non-protease related terminomics experiment (Experimentally Observed). (C) Matrix of substrates and proteases indicating cleavage of substrates. Fields are red where there is cleavage and black where there is none known between the protease and the substrate at this position. The y-axis shows the protein identifier and the position of each terminus. (D) Barplot showing the number of cleavages of each protease in the list. Bars of proteases whose cleavages are enriched in the list are in red, others in blue. (E) IceLogo of the sequences in the list.
