Figure 3.
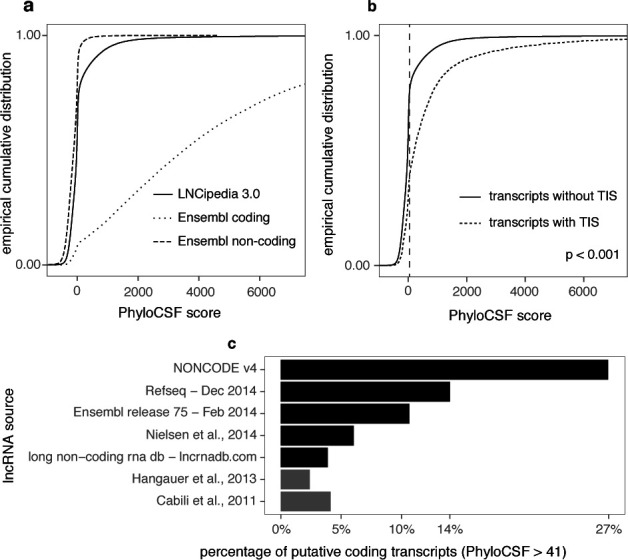
Different methods suggest contamination of coding sequences in lncRNA data sets. (a) PhyloCSF benchmarking and score distributions. We can observe a considerable difference between the score distributions of coding and non-coding transcripts in the Ensembl data set. In addition, while the great majority of LNCipedia is presumably non-coding, it also contains a fraction of transcripts with a PhyloCSF score in the coding range. (b) Transcripts with a TIS have a significantly higher PhyloCSF score (Mann–Whitney U test) compared to other transcripts. (c) Several public lncRNA resources suffer from considerable contamination with protein-coding sequences. The percentage of transcripts with PhyloCSF score greater than 41 is shown for the different sources in LNCipedia 3.0. Two sources already filtered with PhyloCSF are depicted in gray. In the case of RefSeq, only entries with property “biomol_ncrna_lncrna” were considered.
