Figure 2.
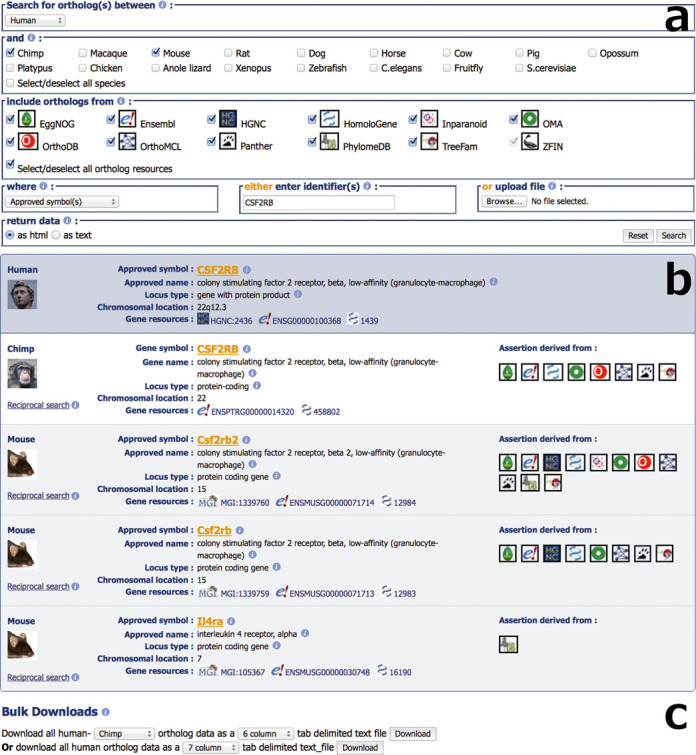
Section ‘a’ shows the updated HCOP search form. Users select a primary species and one or more species that they wish to identify orthologs in. They then select the ortholog resources they wish to include in the search, and the type of search term, e.g. approved symbol, or database identifier, that they are providing. A single search term or list of search terms may be pasted into a text box, or uploaded as a file to be used to run the search. Users can optionally see the results in HTML or plain text format. Section ‘b’ displays an example result panel from HCOP. In this case orthologs between the human gene CSF2RB in chimp and mouse were requested. Information about the query gene appears in the blue section at the top of the results panel, with each ortholog identified having its own section below this. For both the query and ortholog genes we provide basic information about the gene as well as links to the gene in other resources. Each ortholog section has an additional column labelled ‘Assertion derived from’ that contains a set of icons that represent the orthology sources that support this assignment. Section ‘c’ is the bulk downloads section of the HCOP form. This allows users to download pre-computed files of HCOP data from our FTP site, providing fast access to all the data stored in HCOP.
