Figure 1.
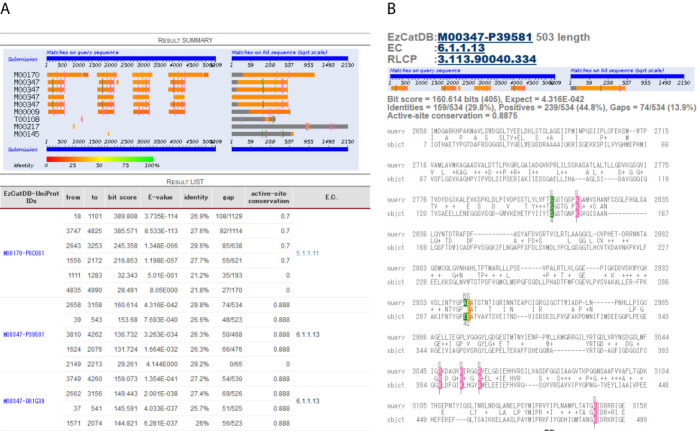
Output for an NRPS sequence (UniProtKB; B8NXQ7) by EzCat-BLAST. (A) Top page. In the ‘RESULT SUMMARY’, sequence matches between query and hit sequences are shown as colored bars along with active-site matches. For this query sequence, four adenylation domains of NRPS, which are homologous to the representative sequences from EzCatDB entries, M00170, M00347 and M00009, were detected. In the ‘RESULT LIST’, the corresponding EzCatDB and UniProtKB IDs for the top hit sequences are shown with the position of the query sequence, bit score, E-value and active-site conservation for each hit sequence. (B) Alignment display for the query and hit sequences. Active-site residues are colored according to the type of active-site residues. Sidechain catalytic residues are presented in magenta, whereas mainchain catalytic residues are shown in green. The cofactor binding residue is shown in orange. The link to EzCatDB entry is also shown with the alignment data.
