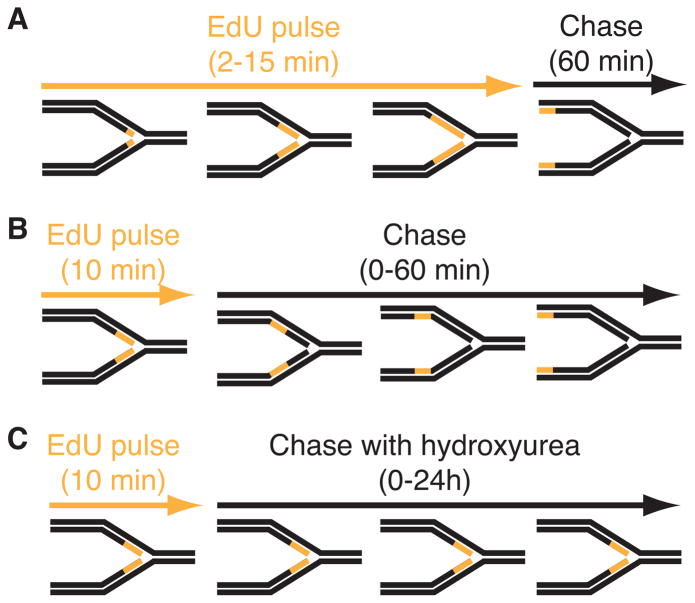
Figure 1.
Diagram of three types of iPOND experiments. In these schematics, the black lines represent unlabeled DNA and orange lines represent DNA labeled with EdU. (A) Experiment to detect proteins that localize at elongating replication forks. Cells are incubated for increasing times in EdU prior to fixation. A single chase sample in which the EdU is removed for one hour prior to fixation is included as a control. True replication proteins will no longer be enriched in this chase control. (B) Experiment to monitor chromatin deposition and maturation. A single short EdU labeling period is followed by increasing chase times to monitor how the proteins associated with the EdU-labeled DNA fragment change as a function of time and distance from the fork. (C) Experiment to assess replication stress responses. Cells are labeled with EdU and then chased into media containing a replication stress-inducing drug like hydroxyurea. Variants of this procedure in which the replication stress agent is added prior to EdU or EdU remains in the growth media during the replication stress period are possible. All iPOND experiments should include an additional control sample that either lacks EdU labeling or in which the biotin-azide is omitted from the procedure (a “no-click” control).