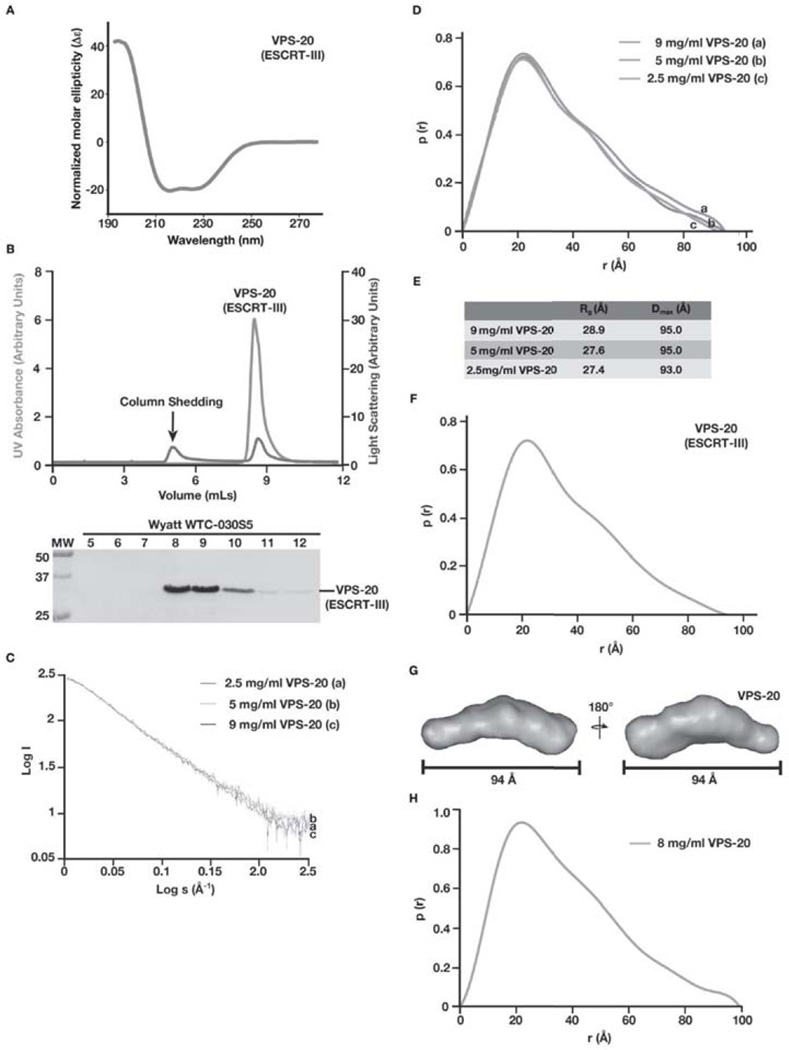
Figure 3. C. elegans VPS-20 exhibits an extended, open conformation in solution.
(A) Circular dichroism (CD) spectroscopy was used to characterize VPS-20. Samples were analyzed at a concentration of 1 µM, and the data were normalized. CD spectra were collected at 25°C in 25 mM sodium phosphate (pH 7.2) using a 1 mm path length quartz cell. (B) Purified, untagged VPS-20 was separated over a gel filtration column that was coupled to a multi-angle light scattering device. Both the UV absorbance (green) and light scattering (blue) profiles are plotted (top) and eluted fractions were separated by SDS-PAGE and stained using Coomassie to highlight the elution profile of VPS-20 (bottom). A small light scattering peak, which contains no protein, is indicated (arrow), and is a result of non-specific column shedding. (C) Log of scattered intensity vs. log of s is shown across the three concentrations of VPS-20 used in SAXS experiments (9 mg/mL, 5 mg/mL, and 2.5 mg/mL). Plots for each concentration of protein are labeled alphabetically (2.5 mg/mL is denoted ‘a’, 5 mg/mL is denoted ‘b’, and 9 mg/mL is denoted with ‘c’). (D) Pair distance distribution function plots comparing the three concentrations of VPS-20 used in SAXS experiments. Plots for each concentration of protein are labeled alphabetically (9 mg/mL is denoted ‘a’, 5 mg/mL is denoted ‘b’, and 2.5 mg/mL is denoted with ‘c’). (E) A summary of the Rg and Dmax values for the three concentrations of C. elegans VPS-20 tested. (F) Pair distance distribution function plot of the merged data set generated from two different SAXS data sets (5 mg/mL and 2.5 mg/mL) determined a Dmax of 94 Å for VPS-20. (G) A low resolution, ab initio model of VPS-20 was determined based on fifteen structures that were generated using the program DAMMIF and averaged with DAMAVER, resulting in a NSD value of 0.69. Two views, rotated 180° related to one another, are shown. (H) Pair distance distribution function plot of VPS-20 (8 mg/mL), as measured by SAXS in the absence of salt.