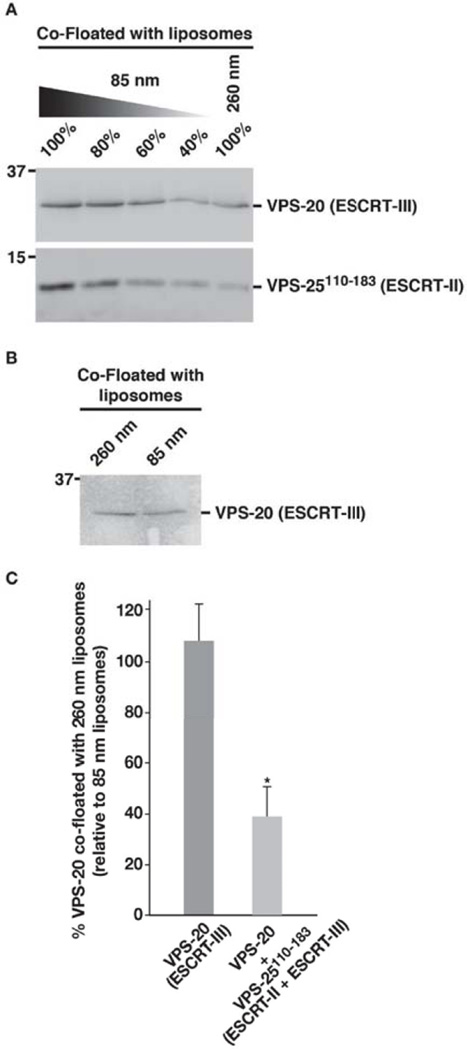
Figure 7. A fragment of VPS-25 is sufficient to enable VPS-20 to sense membrane curvature.
(A) A co-flotation assay was used to analyze the binding of a mixture of VPS-20 and VPS-25110–183 (amino acids 110–183) to liposomes of different diameters (85 and 260 nm). Fractions were resolved by SDS-PAGE and immunoblotted using VPS-20 or ESCRT-II antibodies. A dilution series of the protein mixture that co-floated with 85 nm liposomes was loaded to quantify the relative amount that co-floated with 260 nm vesicles. Data shown are representative of at least three independent experiments. On the basis of densitometry measurements, we found that less than 50% of the protein mixture co-floated with 260 nm vesicles, relative to 85 nm vesicles. (B) A co-flotation assay performed similarly as described in panel A demonstrates that VPS-20 alone fails to sense the difference in curvature between 85 and 260 nm liposomes. (C) Quantification of the co-flotation assays performed in panels A and B. The percentage of VPS-20 co-floated with 260 nm vesicles relative to that co-floated with 85 nm vesicles is plotted for each condition shown. Error bars represent mean +/− S.E.M. (n=3 each; p < 0.05, t-test).