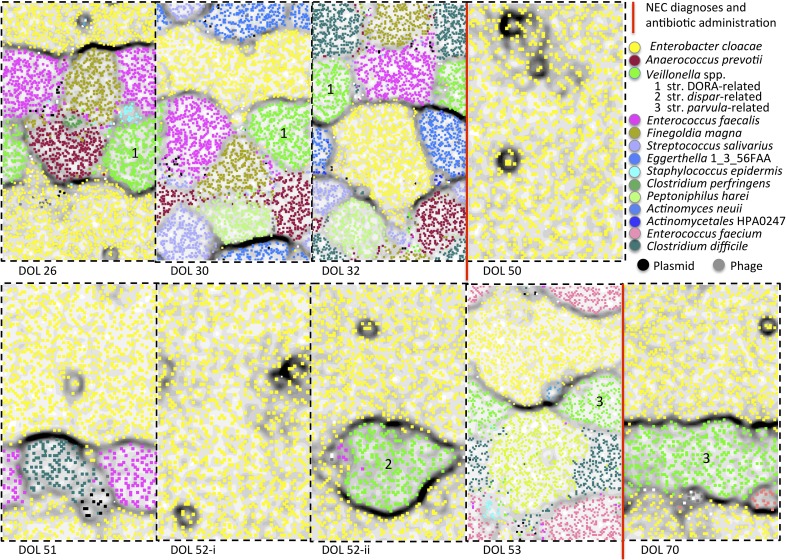
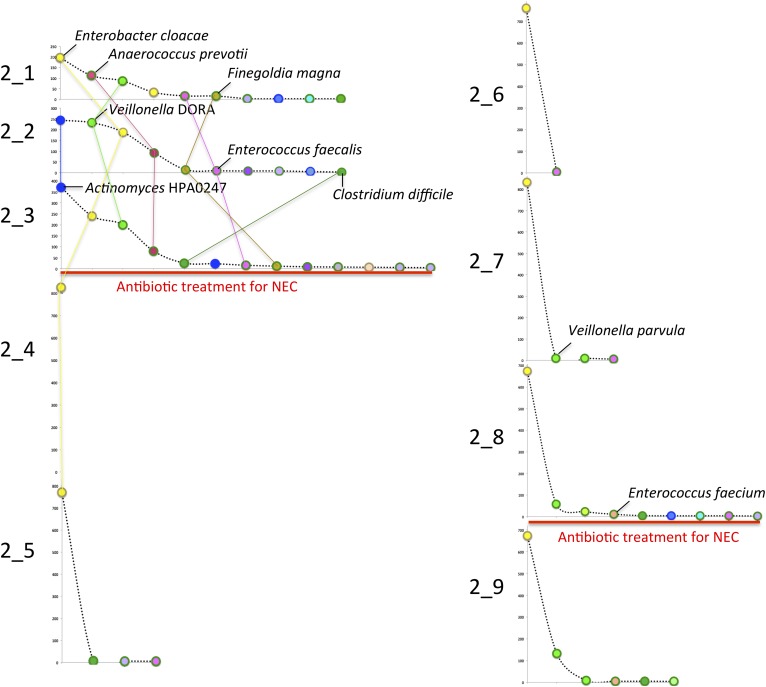
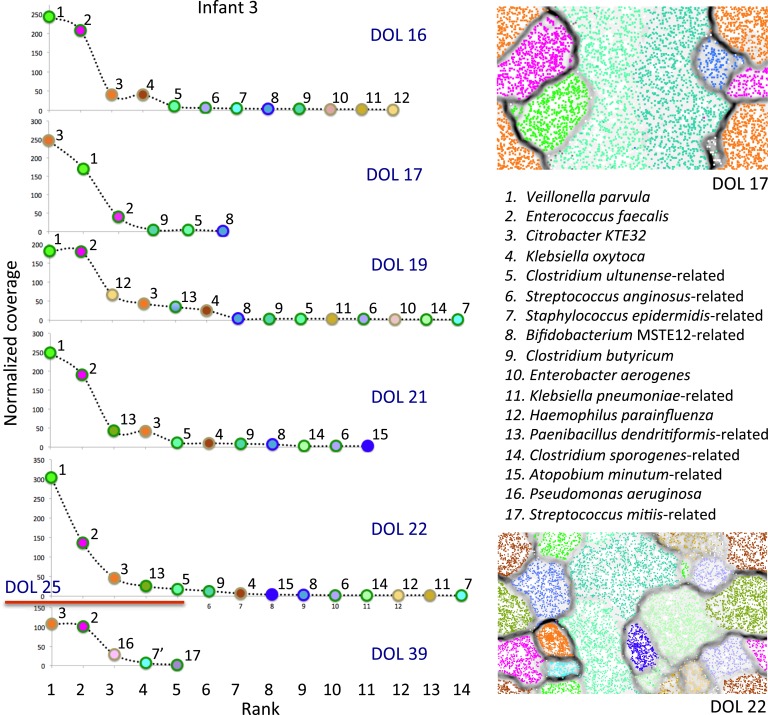
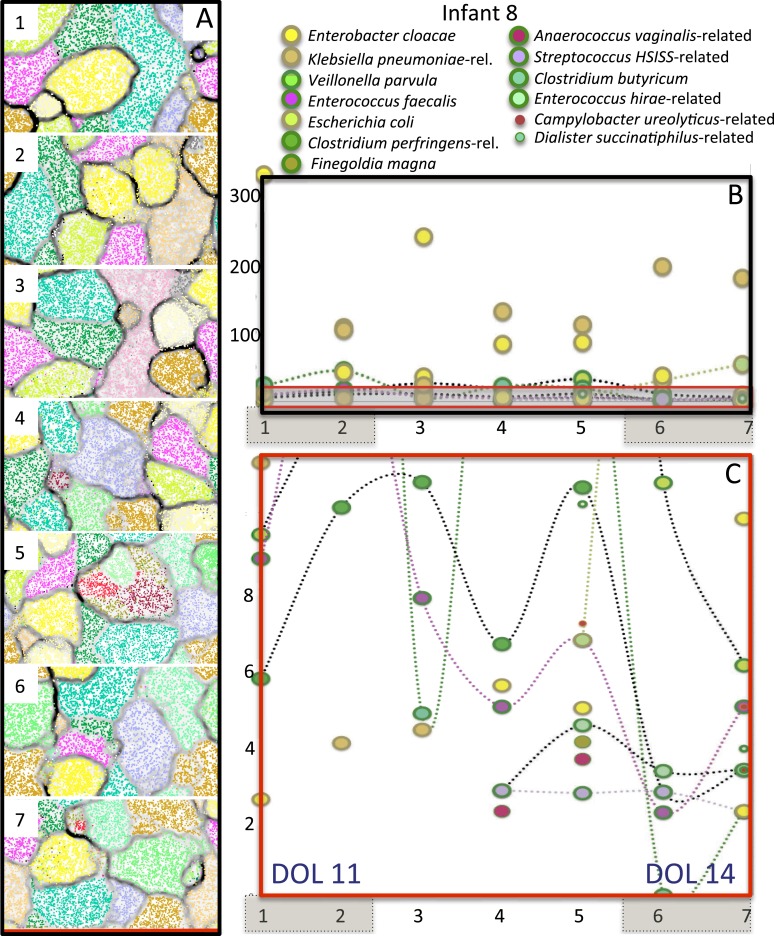
Figure 8. Microbial community composition, community complexity, and an overview of binning for samples from infant #2.
The diagrams are unit repeats of a tetranucleotide emergent self organizing map; points coded to reflect the bin assignment of the scaffold verify the binning (see ‘Materials and methods’ section). Vertical red lines separate samples before and after antibiotic administration to treat necrotizing enterocolitis (NEC) (two instances). Organisms are listed primarily in order of abundance in the first sample. Note that, with the exception of the dominant member, Enterobacter cloacae, species representation changed dramatically following antibiotic administration. The Veillonella strain varied (numbers differentiate areas that represent different populations).