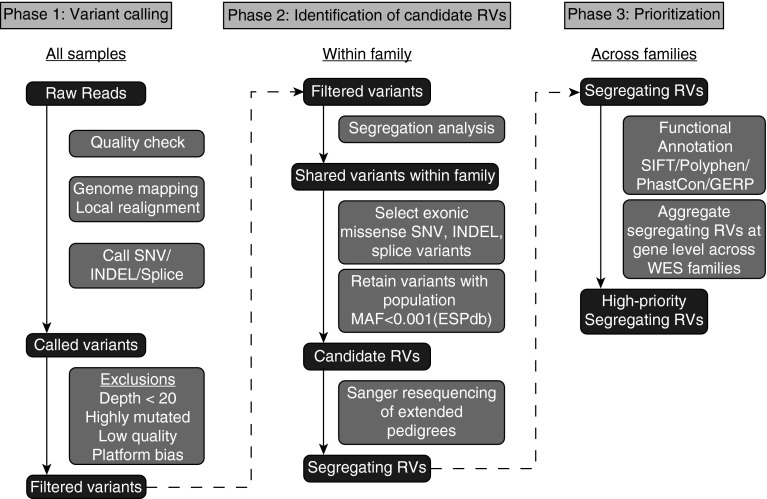
Figure 1.
Whole-exome sequencing (WES) analysis pipeline. WES analysis occurred through three phases. In phase 1, raw reads from all samples underwent quality checking, genome mapping, local realignment, and variant calling using Varscan 2.0 to call variants. Following exclusionary quality-control filter, in phase 2 only variants that were found in all affected WES subjects within a family were retained. From among these segregating variants, only those highly likely to affect protein quantity or function (nonsynonymous exonic single-nucleotide variants [i.e., missense or nonsense], insertions and deletions, splice variants) were selected for further analysis. From among these variants, those with minor allele frequency (MAF) greater than 0.001 in the Exome Sequencing Project database (ESPdb) were excluded to yield candidate rare variants (RVs). Sanger requencing was then performed in extended pedigrees encompassing all affected subjects for whom DNA was available to exclude variants that did not segregate fully with disease. These segregating RVs from each family were functionally annotated and aggregated at the gene level across families for further study. GERP = genomic evolutionary rate profiling; INDEL = insertion and deletion; SIFT = Sorting Intolerant from Tolerant; SNV = single-nucleotide variants.