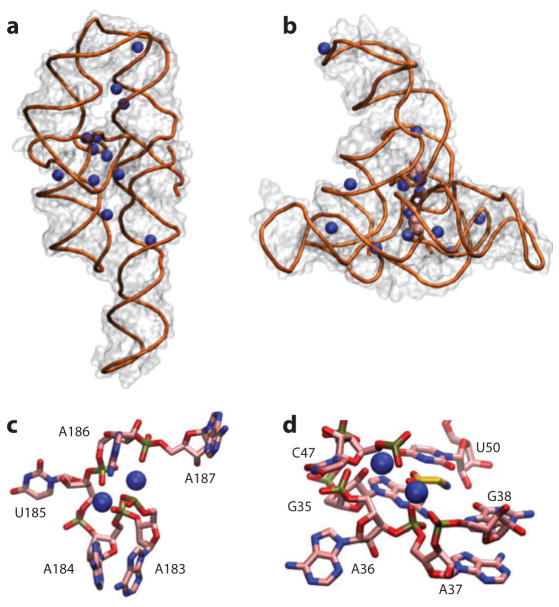
Figure 2.
Crystal structures reveal details of ion–nucleic acid interactions but fail to give a complete picture of the ion atmosphere. (a) Crystal structure of the P4–P6 domain of the Tetrahymena group I intron ribozyme [Protein Data Bank (PDB) identifier 1GID] (21). Twelve crystallographically resolved Mg2+ ions are shown as blue spheres. (b) Crystal structure of the tandem aptamer of a glycine riboswitch from Fusobacterium nucleatum (PDB 3P49) (195). The two glycine molecules bound to the tandem aptamer are shown in pink. Thirteen crystallographically resolved Mg2+ ions are shown as blue spheres. (c) Close-up of the metal ion core of the P4–P6 RNA. Metal ions (spheres) interact with the adenine (A)-rich bulge, making a number of contacts and organizing the RNA residues. (d) Detailed rendering of the glycine binding pocket of aptamer I of the structure in panel b. The two crystallographically resolved Mg2+ ions (spheres) coordinate a series of backbone residues in the binding pocket as well as the ligand glycine. These RNAs have 158 and 169 residues, corresponding to total charges of the RNAs of −157 and −168, respectively. The structures resolve 12 and 13 Mg2+, respectively, which means that only ~15% of the ion atmosphere required for charge neutrality is accounted for by the explicitly resolved ions. In addition, the crystallographically resolved ions can be replaced by ions of other identities; both RNAs can undergo partial folding in high concentrations of monovalent ions (62, 83). Docking of the P5abc metal ion core in the P4–P6 RNA (c) and glycine binding (d) and folding to the native state for the tandem aptamer riboswitch require divalent ions, however, with limited specificity between different divalent species (52, 62, 103).