Figure 1.
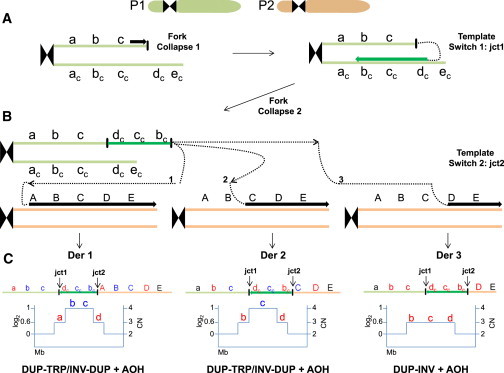
Replication-Based Mechanism Model for Generation of DUP-TRP/INV-DUP Rearrangements followed by AOH
(A) Event described probably occurred in a post-zygotic mitotic cell involving parental homolog chromosomes, P1 and P2. It might initiate due to a stalled or collapsed replication fork that uses a complementary strand to resume replication (template switch 1), generating jct1. Microhomology in the complementary strand at the annealing site (dc) is used to prime DNA synthesis, which will establish a unidirectional replication and will produce an inverted-oriented segment as compared to the reference genome.
(B) A new fork stalling or collapsing event releases a free 3′ end that can be resolved by a new cycle of template switching (template switch 2) and target annealing, this time using the homolog chromosome to prime and resume DNA synthesis. This second event can generate jct2 as well as AOH. It is possible that multiple cycles of fork collapse and target annealing occurs and produces additional complexities.
(C) Top: different genomic structures (Derivative [Der] 1, 2, 3) are expected to be generated depending on the location of the selected annealing site to prime DNA synthesis in the second template switch event. For instance, if the new annealing occurs at or before allele A to C, a triplication flanked by duplications will be produced (DER 1 and DER 2), whereas if annealing occurs at allele D, an inverted duplication will result (DER 3). Annealing at allele E will produce an inversion of segments b-c-d along with a partial duplication of b-c (not shown). AOH will result if unidirectional replication fork continues till the telomere. Bottom: expected segmental copy number (CN) variation in a simulated aCGH experiment. a, b, c: representative chromosome alleles; ac, bc, cc: complementary chromosome alleles; A, B, C, D, E: corresponding homolog chromosome alleles.
