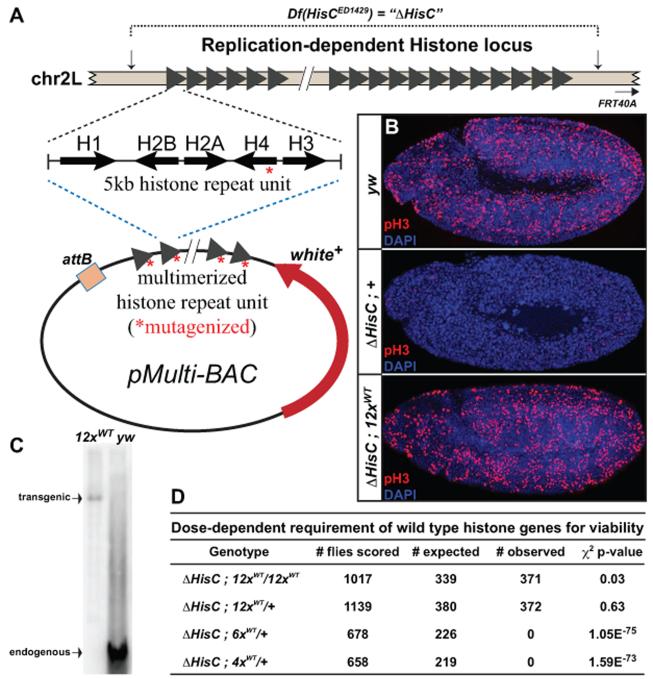
Figure 1. A single histone replacement transgene rescues deletion of the endogenous histone locus.
(A) Schematic of the endogenous replication-dependent histone locus. Breakpoints of the deficiency are indicated by vertical arrows. Each triangle represents a single 5kb histone repeat unit, which was cloned and multimerized into a BAC vector for transgenesis. MCS: Multiple Cloning Site; w+: mini-white cassette; attB: site-specific recombination sequence. (B) Confocal images of cycle 15 embryos stained with antibodies for phospho-histone H3 (red) and DAPI (blue). Genotypes (top to bottom): wild type (yw); homozygous histone deletion (ΔHisC;+); homozygous histone deletion with two copies of the 12× wild type histone transgene (ΔHisC;12×WT). (C) Southern blot of SalI/XhoI digested genomic DNA from 12×WT and wild type (yw) flies, hybridized with an H2A probe. (D) Table of viability tests for various wild type histone arrays. The p-value for the chi-square test is shown. See also Figure S1.