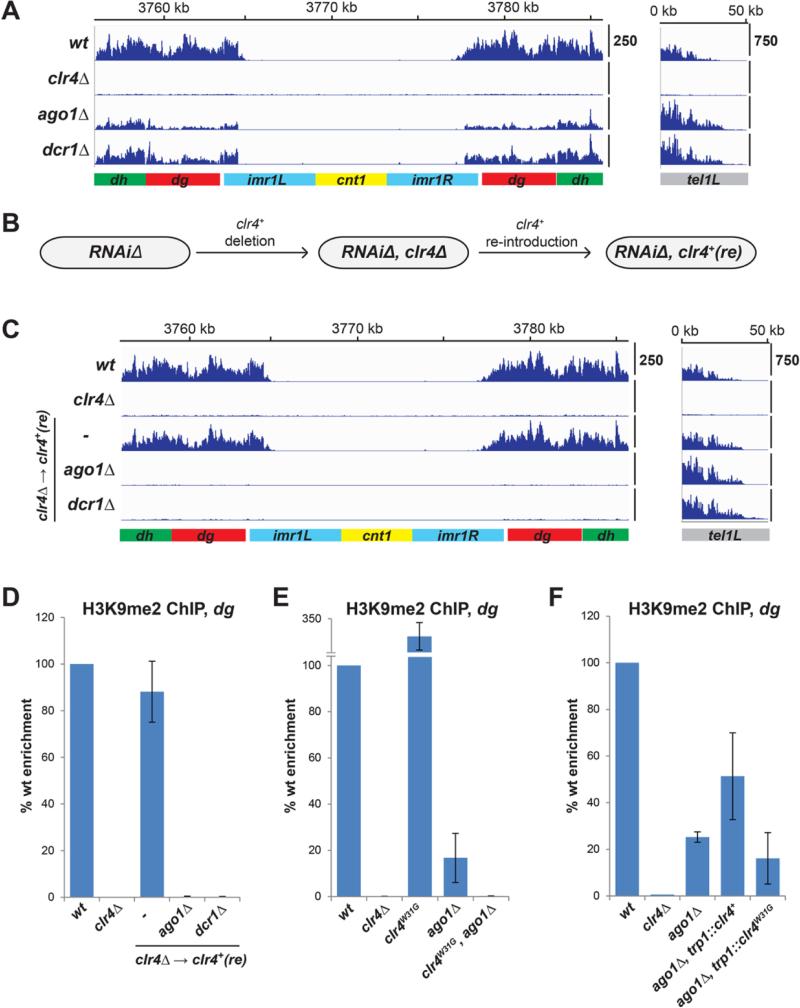
Fig. 7. RNAi-independent H3K9 methylation at pericentromeric repeats is epigenetically inherited.
(A) ChlP-seq experiments showing the persistence of residual histone H3K9me2 at the pericentromeric dg and dh repeats of chromosome 1 in agolΔ and dcrlΔ cells. Libraries were sequenced on the lllumina HiSeq2500 platform and normalized to read per million (y axis). Chromosome coordinates are indicated above the plots. (B) Scheme for the reintroduction of clr4+ into RNAiΔ, clr4Δ cells to test the requirement for RNAi in H3K9me establishment. clr4+ was reintroduced to the native locus to avoid overexpression. (C) ChlP-seq experiments showing that the re-introduction of clr4+ into clr4Δ cells, but not clr4Δ agolA or clr4Δ dcrlΔ cells, restores H3K9me2 at the pericentromeric repeats of chromosome 1 (left). Reads for H3K9me2 at the telomeres of chromosome 1 (telLl) on the right side show that, unlike the centromeres, establishment of telomeric H3K9me does not require RNAi. (D) ChlP-qPCR experiments verify that RNAi is required for the reestablishment of H3K9me2 at the pericentromeric dg repeats. (E) ChlP-qPCR experiments show that a mutation in the chromodomain of Clr4 (clr4W31G) abolishes the maintenance of H3K9me2 at dg repeats. (F) ChlP-qPCR experiments show that an additional copy of wild type clr4+, but not clr4W31G, boosts residual H3K9me2 levels at dg. Error bars represent standard deviations.