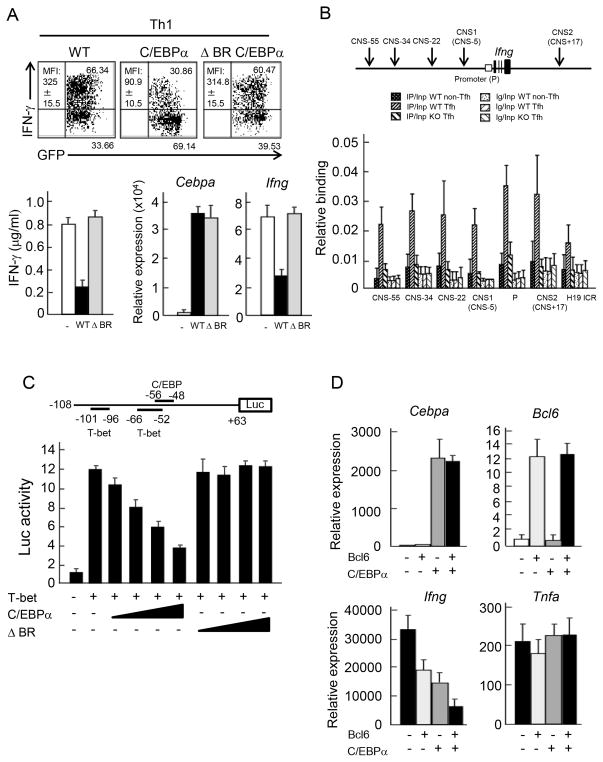
Figure 7. Inhibition of T-bet-mediated IFN-γ transcription by C/EBPα.
(A) Naive CD4+ T cells derived from OT-II TCR Tg mice were transduced with retroviruses carrying pGFP-RV(−), pGFP-RV- Cebpa (C/EBPα) and pGFP-RV-ΔBR Cebpa (ΔBR C/EBPα) with stimulation by peptide-loaded APCs under Th1 skewing culture. After 4 days, IFN-γ expression was analyzed in infected T cells by ICS (upper) and by ELISA (lower left). MFI of IFN-γ expression in infected cells was indicated. mRNA expression of Cebpa and Ifng was measured by quantitative PCR (lower right). Data is a representative of three independent experiments with similar results. (B) Upper panel indicates a schematic diagram of the Ifng locus. C/EBPα binding sites were accessed by ChIP analysis. PCR amplification was conducted with DNA prepared from chromatin precipitated with control Ig and anti-C/EBPα Abs in non-Tfh and Tfh cells isolated from immunized WT or Cebpa KO mice by FACS sorting. Primer set for H19 ICR locus was used as negative control. Data is a representative of two independent experiments with similar results. (C) A schematic diagram of the luciferase Ifng promoter reporter constructs (upper). +63 to −108 region from transcription start site in Ifng gene was used as a promoter. −56 to −48 region contains a C/EBP binding site. −66 to −52 and −101 to −96 regions shown contain T-box protein binding sites. Luciferase assay of 293T cell transduced with pGFP-RV(−), pGFP-RV-Cebpa (C/EBPα) or pGFP-RV-ΔBR Cebpa (ΔBR C/EBPα) together with pGFP-RV-T-bet was performed (lower). Data is a representative of three independent experiments with similar results. (D) Naive CD4+ T cells derived from OT-II TCR Tg mice were stimulated with peptide loaded APCs under Th1 skewing culture and further infected with combination of retroviruses carrying pGFP-RV(−), pGFP-RV-Bcl6 (Bcl6) pMIGRII-RV(−) and pMIGRII-RV-Cebpa (C/EBPα) as indicated. After 4 days, GFP+ hCD2+ cells were isolated and re-stimulated with anti-CD3 Ab. Then, mRNA expression of Cebpa, Bcl6, Ifng and Tnfa was measured by quantitative PCR. Data are a representative of two independent experiments.