Abstract
DNA sequencing and analysis of two distinct C—P lyase operons in Pseudomonas stutzeri WM88 were completed. The htxABCDEFGHIJKLMN operon encodes a hypophosphite-2-oxoglutarate dioxygenase (HtxA), whereas the predicted amino acid sequences of HtxB to HtxN are each homologous to the components of the Escherichia coli phn operon, which encodes C—P lyase, although homologs of E. coli phnF and phnO are absent. The genes in the htx operon are cotranscribed based on gene organization, and the presence of the intergenic sequences is verified by reverse transcription-PCR with total RNA. Deletion of the htx locus does not affect the ability of P. stutzeri to grow on phosphonates, indicating the presence of an additional C—P lyase pathway in this organism. To identify the genes comprising this pathway, a Δhtx strain was mutagenized and one mutant lacking the ability to grow on methylphosphonate as the sole P source was isolated. A ca.-10.6-kbp region surrounding the transposon insertion site of this mutant was sequenced, revealing 13 open reading frames, designated phnCDEFGHIJKLMNP, which were homologous to the E. coli phn genes. Deletion of both the htx and phn operons of P. stutzeri abolishes all growth on methylphosphonate and aminoethylphosphonate. Both operons individually support growth on methylphosphonate; however, the phn operon supports growth on aminoethylphosphonate and phosphite, as well. The substrate ranges of both C—P lyases are limited, as growth on other phosphonate compounds, including glyphosate and phenylphosphonate, was not observed.
The essential nutrient phosphorus is widely held to be a redox conservative element in living systems, where it occurs only in the +5 valence state of inorganic phosphate and its organic esters, amides, and anhydrides. Nevertheless, many microorganisms are capable of metabolizing compounds containing P at lower redox states, including hypophosphite (8, 9, 22), phosphite (1, 3, 7, 8, 15, 22, 28), and phosphonic acids (for a review, see reference 30). Phosphonates, in particular, are known to be ubiquitous in nature, and in some ecosystems they comprise a major fraction of the available P (5, 10, 13). These compounds are characterized by very stable C—P bonds, in contrast to the labile C—O—P bonds found in phosphate esters. Phosphonates are produced by a variety of organisms, including both prokaryotes and eukaryotes, and can be found in the form of phosphonolipids and as side groups on polysaccharides and glycoproteins (10). In addition, a wide variety of phosphonate antibiotics are produced by microorganisms, mostly by members of the genus Streptomyces (29).
Given the prevalence of phosphonates in nature, it is not surprising that microorganisms have also evolved with the capacity to consume these compounds. A variety of pathways that allow specific phosphonates to be used as either the sole P or the sole carbon source have been discovered (30). The most widespread of these pathways involves the enzyme C—P lyase, which allows a variety of phosphonates to be used as sole P sources. As its name implies, C—P lyase is believed to catalyze the direct cleavage of carbon—phosphorus bonds to produce the corresponding alkane and phosphate. There is some question as to the actual products of the reaction, however, because in vitro activity has never been observed for this enzyme, despite the efforts of several research groups (4, 16, 17, 31). Thus, its products have been inferred from studies of whole or permeabilized cells.
In contrast to the dearth of biochemical data, genetic studies of Escherichia coli have provided considerable insight into the metabolism of P compounds by the C—P lyase pathway. A series of genetic studies revealed that a fourteen-gene operon, phnCDEFGHIJKLMNOP, encodes proteins required for the uptake and assimilation of phosphonates via a C—P lyase pathway (4, 19-21, 33). PhnCDE comprises a binding protein-dependent transporter with the capacity to transport not only phosphonates but also phosphate esters, phosphite, and phosphate. PhnG, PhnH, PhnI, PhnJ, PhnK, PhnL, PhnM, and PhnP are thought to comprise a multisubunit C—P lyase or, alternatively, enzymes in a multistep pathway for C—P bond cleavage. PhnF and PhnO are not required for phosphonate degradation, but are likely to be transcriptional regulators. The role of PhnN remains obscure. Although PhnN is not required for phosphonate degradation, it does appear to be involved, because phnN mutants grow poorly on media with phosphonates as the sole P source. PhnN was recently shown to catalyze the formation of ribose-phosphate esters (11), leading to the suggestion that the natural function of PhnN may be to produce a ribose-phosphonate ester, which may be an intermediate in phosphonate catabolism.
Surprisingly, the examination of E. coli phn operon mutants also revealed that these genes were required for the utilization of phosphite (P valence, +3) as the sole P source (20). Thus, in addition to its role in C—P bond cleavage, C—P lyase can also oxidize phosphite to phosphate. (The use of any P compound as the sole P source requires its conversion to phosphate, because phosphate is required for innumerable cellular processes.) Evidence of the genetic linkage between the metabolisms of different reduced P compounds was further strengthened by recent studies of the utilization of hypophosphite as the sole P source by Pseudomonas stutzeri.
Genetic and biochemical studies have shown that in P. stutzeri, hypophosphite is oxidized to phosphate via a phosphite intermediate. These reactions are catalyzed by two novel enzymes, hypophosphite-2-oxoglutarate dioxygenase and phosphite:NAD oxidoreductase (6, 22, 34), which are encoded by discrete genetic loci located ca. 15 kbp apart on the P. stutzeri chromosome. Phosphite oxidation in P. stutzeri was demonstrated to be primarily a function of the ptxABCDE locus, which, in addition to NAD:phosphite oxidoreductase (ptxD), encodes a putative binding protein-dependent phosphite transporter (ptxABC) and a putative transcriptional regulator (ptxE) (22). Partial sequencing of the htx locus, which encodes hypophosphite-2-oxoglutarate dioxygenase (HtxA), revealed nine open reading frames (ORFs), designated htxABCDEFGHI′. While htxA was required for hypophosphite oxidation in P. stutzeri, htxBCDEFGHI′ was not. Interestingly, these genes are highly homologous to phnCDEFGHIJ of E. coli.
Given the ability of C—P lyase to oxidize phosphite to phosphate, it seemed possible that the htx operon could encode a complete pathway for the oxidation of hypophosphite to phosphate in a manner independent of the PtxD pathway. Thus, HtxA would oxidize hypophosphite to phosphite, and the putative htx-encoded C—P lyase would oxidize phosphite to phosphate. The observation that a Δptx mutant shows low levels of growth on phosphite as the sole P source after prolonged incubation is consistent with the existence of an additional phosphite oxidation pathway (A. K. White and W. W. Metcalf, unpublished results). We suspected that a complete C—P lyase operon may be present within the htx locus of P. stutzeri WM88. However, because the sequence of the htx locus was not complete, this hypothesis could not be verified. Moreover, strains with a deletion of the entire region encompassing both ptx and htx remained capable of growth on phosphonates, suggesting the existence of an additional, unidentified C—P lyase encoded elsewhere on the chromosome.
In this report, we show that htx does indeed encode a functional C—P lyase and that an additional and functional C—P lyase is present in P. stutzeri. Further, genetic analysis of mutants with defined deletions of two C—P lyase operons and of the ptx operon shows that C—P lyase can indeed play a role in phosphite and hypophosphite oxidation but not in the manner that was initially hypothesized.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
All P. stutzeri strains described are derivatives of the spontaneous streptomycin-resistant, smooth colony form of the original strain, P. stutzeri WM567, which in turn is a derivative of the original hypophosphite-oxidizing isolate P. stutzeri WM88 (22). Cloning experiments were performed with either DH5α/λpir (23) or BW20767 (18). BW20767 was also used as a donor strain in conjugation experiments. The suicide plasmid pAW19 is a derivative of pWM91 (18), which contains the BamHI kanamycin resistance cassette from pUC4K. Plasmid pWM234 (22) was used as a template for sequencing the remainder of the htx locus. For most experiments, Luria-Bertani broth or tryptone-yeast extract (TYE) agar plates containing the appropriate antibiotic were used (32). Kanamycin was added at 50 μg/ml, and streptomycin was added at 100 μg/ml. Selection of transformants for cloning experiments with pAW19 was done on TYE agar containing kanamycin. Selection for exconjugants harboring integrated pAW19 derivatives was done on 0.2% glucose MOPS (morpholinepropanesulfonic acid) minimal medium containing kanamycin. Sucrose-resistant recombinants resulting from segregation of pAW19 derivatives were counterselected on TYE agar with 50 g of sucrose replacing the NaCl (32).
P oxidation phenotypes were scored at 37°C on 0.2% glucose MOPS agar containing a 0.5 mM concentration of the appropriate P source. All P compounds used in this study were purchased from Sigma (St. Louis, Mo.). The purity of some P compounds with respect to contaminating P-containing compounds was assessed by 31P nuclear magnetic resonance as described in reference 34. Compounds that failed to support growth were assumed to be free of contaminating levels of phosphate. All compounds were greater than 99% pure with respect to their P content. When glyphosate (phosphonomethylglycine) was used as the P source, the media were supplemented with 0.05 mg of an aromatic-amino-acid mixture/ml. To remove the contaminating phosphate, the agar and all glassware were rinsed with multiple changes of ultrapure deionized water prior to use. The solutions of all P compounds were made immediately prior to use and were filter sterilized.
DNA methods.
Standard methods were used for the isolation and manipulation of plasmid DNA. Chromosomal DNA was isolated from P. stutzeri strains as described previously (2). DNA hybridizations were carried out by using a digoxigenin system as recommended by the manufacturer (Roche, Mannheim, Germany). Sequencing reactions were performed with the BigDye sequencing reagent (Applied Biosystems, Foster City, Calif.) as recommended by the manufacturer and were analyzed at the W. M. Keck Center for Comparative and Functional Genomics at the University of Illinois at Urbana.
Plasmid construction.
Plasmid pJB1 carries a deletion of htxA-htxO in a pAW19 vector backbone. To create the deletion, ca. 1.0 kbp of sequence upstream of the htxA translational start site and ca. 1.0 kbp of sequence downstream of htxO were amplified by PCR using Taq DNA polymerase and the primers listed in Table 1. The PCR products were digested with the appropriate restriction enzymes and ligated to the SpeI and SacI sites of pAW19.
TABLE 1.
Oligonucleotide primers used for deletion constructions
| Deletion | Sequence amplified | Primer seta | Restriction enzyme site |
|---|---|---|---|
| Δhtx | Upstream htxA | 5′-GGCGGCGGCACTAGTCTGCCCCGATATCAAGCAAC-3′ | SpeI |
| 5′-GGCGGCGGCGCGGCCGCGTGATGCTCCAAGGTCTTCC-3′ | NotI | ||
| Downstream htxO | 5′-GGCGGCGGCGCGGCCGCATGGCATCGAGTGCTCAAC-3′ | NotI | |
| 5′-GGCGGCGGCGAGCTCGCCGGACATTTGTATACGC-3′ | SacI | ||
| Δptx | Upstream ptxA | 5′-GGCGGCGGCACTAGTGGCTAGCATCACACAGAAACC-3′ | SpeI |
| 5′-GGCGGCGGCGCGGCCGCCCTTGTTACCGCACTGCTTC-3′ | NotI | ||
| Downstream ptxE | 5′-GGCGGCGCGGCCGCGGTGATGGATGGTTGCGAT-3′ | NotI | |
| 5′-GGCGGCGAGCTCCAGCGTGGCGTAGAGCTGCG-3′ | SacI | ||
| Δphn | Upstream phnC | 5′-GGCGCGCCGAGCTCACCAGGCCTGGCGTGTCGGC-3′ | SacI |
| 5′-GGCGCGCCGCGGCCGCTCTGCGCGGCCGGGGAGGT-3′ | NotI | ||
| Downstream phnP | 5′-GGCGCGCCGCGGCCGCCGCTTAGGAGAGAAGCCGAG-3′ | NotI | |
| 5′-GGCGCGCCAACTAGTGTTCGGGCAAGCGCTCCAGC-3′ | SpeI |
The sequences of the restriction sites incorporated into the PCR primers are underlined.
Plasmid pJB2 carries a complete deletion of both the ptx and htx operons (ptxA-orf344) in a pAW19 vector backbone. To create this deletion, ca. 1.0 kbp of sequence directly upstream of the ptxA translational start site was amplified by PCR with Taq DNA polymerase and the primers listed in Table 1. The resulting PCR fragment was digested with SpeI and NotI and was inserted into the same sites of pJB1, resulting in the replacement of the htxA upstream fragment with the ptxA upstream fragment.
Plasmid pAW52 carries a phnC-phnP deletion in a pAW19 vector backbone. To construct the deletion, ca. 1.0 kbp of sequence upstream of the phnC translational start site and 1.0 kbp of sequence directly downstream of the phnP stop codon were amplified by PCR with Pfu Turbo DNA polymerase and the primers listed in Table 1. The resulting PCR products were digested with the appropriate restriction enzymes and were inserted between the SpeI and SacI sites of pAW19.
Plasmid pAW79 carries a ptxA-ptxE deletion. To construct this deletion, the 1.0 kbp of sequence upstream of the ptxA translational start site and the 1.0 kbp of sequence downstream of the ptxE stop codon were amplified by PCR with Pfu Turbo DNA polymerase and the primers listed in Table 1. The resulting PCR products were digested with the appropriate restriction enzymes and were inserted between the SpeI and SacI sites of pAW19.
Genetic techniques.
The in vitro-constructed deletions were recombined onto the chromosome of P. stutzeri WM567 by sacB counterselection as previously described (18). The conjugative transfer of plasmids from E. coli BW20767 to P. stutzeri WM567 was done by using the filter mating technique as previously described (14), with selection of exconjugants on 0.2% glucose MOPS medium containing 0.5 mM phosphate and kanamycin. Sucrose-resistant segregants were screened for loss of the integrated plasmid by scoring their kanamycin sensitivity. The correct mutant constructs were differentiated from wild-type segregants by DNA hybridization analysis using as a probe the same plasmid used to create the deletion.
Isolation and cloning of the phn genes of P. stutzeri was done in a strain of P. stutzeri WM1926 (rpsL ΔptxA-orf344) by in vivo Tn5 mutagenesis with pRL27 as previously described (14). Transposon mutants were screened for the inability to grow on glucose-MOPS medium with methylphosphonate as the sole source of P. The BamHI chromosomal fragment containing the Tn5 insertion site and flanking region was cloned to create plasmid pMW5. This plasmid carries the phnI::Tn5-RL27 insertion along with a substantial region of flanking DNA and was used as a sequencing template for the phn locus of P. stutzeri.
RT-PCR.
Total RNA was isolated from cultures of P. stutzeri grown to mid-logarithmic stage (optical density at 600 nm of ca. 0.6) in 0.2% glucose MOPS minimal medium with 0.5 mM hypophosphite as the sole source of P. RNA isolation was carried out with an RNeasy mini kit with RNAprotect bacterial reagent (QIAGEN, Inc., Valencia, Calif.) per the manufacturer's instructions. To remove contaminating chromosomal DNA, the RNA preparation was digested with amplification grade DNase I (Invitrogen, Carlsbad, Calif.). The DNase I-treated RNA was then used as a template in a reverse transcription (RT) assay using SuperScript II RNase H− reverse transcriptase (Invitrogen), according to the manufacturer's protocol. PCR amplification of the cDNA from the RT reaction was performed with Platinum Pfx DNA polymerase (Invitrogen) as recommended. A positive control, in which only chromosomal DNA was added to the PCR, and a negative control, in which only RNA without reverse transcriptase was used for the PCR, were run under identical PCR amplification conditions. The primers used are listed in Table 2.
TABLE 2.
Oligonucleotide primers used for the amplification of htx junction sequences
| Junction amplified | Primer set | Predicted product size (bp) |
|---|---|---|
| htxAB | 5′-AAGTGTACGACCACCGGAAC-3′ | |
| 586 | ||
| 5′-GTCTGGCTAGGCGTTGGAC-3′ | ||
| htxBC | 5′-TCCTTCGAAGTCCAAGATCG-3′ | |
| 587 | ||
| 5′-CAGCCCCTTTTAGAATGACG-3′ | ||
| htxCD | 5′-CAGCAGGGCAAAGATTATCC-3′ | |
| 560 | ||
| 5′-CCGGAGACAGTTCATGTATGG-3′ | ||
| htxDE | 5′-GTGCATGCGCTTTACCAG-3′ | |
| 583 | ||
| 5′-ACAGCGCAATCTCCAGGTCT-3′ | ||
| htxEF | 5′-AGAACGCCGACAAGAAGC-3′ | |
| 589 | ||
| 5′-TTCGATCGACTGCCCTTG-3′ | ||
| htxFG | 5′-GCATGCCAGGTTGTGAAG-3′ | |
| 573 | ||
| 5′-GCACCTGCTCGTGTTGAATA-3′ | ||
| htxGH | 5′-GGCAACCGTTTGCTGGAG-3′ | |
| 581 | ||
| 5′-CACCGGCACATCCATCAG-3′ | ||
| htxHI | 5′-CCGGTGAGGTACTGATGACC-3′ | |
| 553 | ||
| 5′-AATCAGCGACAGCGTAACCT-3′ | ||
| htxIJ | 5′-CATCAACGGCCACTATGTGA-3′ | |
| 700 | ||
| 5′-CTGGTACACCATGCCAAAGC-3′ | ||
| htxJK | 5′-CCGCTGCTCTACCTCGAC-3′ | |
| 591 | ||
| 5′-ACAGTGCAGAAATTGGGTGA-3′ | ||
| htxKL | 5′-GAACTGCTGGCCCTGCTC-3′ | |
| 556 | ||
| 5′-AACTCCAGCGGGAAGTGC-3′ | ||
| htxLM | 5′-CTGCCTGTGCGGTGACTACT-3′ | |
| 600 | ||
| 5′-GGGCCTCGTCCAGGTATT-3′ | ||
| htxMN | 5′-GAATCGGCATCGAGATCAAC-3′ | |
| 584 | ||
| 5′-CAGCGCAGATGAAAGAGTCC-3′ | ||
| htxN-orf282 | 5′-GCTTCGCCTACCTCACAGAC-3′ | |
| 587 | ||
| 5′-AACTGCGCACCAGAGGATAG-3′ | ||
| orf282-orf344 | 5′-CTCTTGGCGGAACCTGTATC-3′ | |
| 569 | ||
| 5′-GAGCGAATGCCTTTGAGAAC-3′ |
Nucleotide sequence accession numbers.
The GenBank accession numbers for the P. stutzeri WM88 DNA sequences determined in this study are AF061267 for the completed htxIJKLMN and orf282-orf344 sequences and AY505177 for the complete phnCDEFGHIJKLMNP sequence.
RESULTS
Complete sequence analysis of the htx locus.
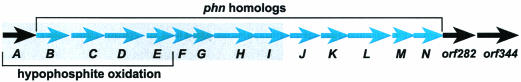
A previously isolated cosmid clone (22) harboring a fragment of P. stutzeri chromosomal DNA containing htxA to htxI and downstream sequence was used as a template to obtain the complete sequence of the htx locus beyond htxI′. Immediately downstream of the nine previously identified ORFs (htxA-htxI) (22), seven additional ORFs were observed. These were designated htxJKLMN, orf282, and orf344 (Fig. 1).
FIG. 1.
Structure of the htx operon of P. stutzeri WM88. Black arrows indicate genes with no homology to phn genes. Blue arrows indicate htx genes that are homologous to phn genes, which comprise all of the components of a complete C—P lyase. The sequences of the genes lying within the shaded gray box had previously been determined (22).
FASTA searches with the predicted amino acid sequences of HtxB to ORF344 (27) were performed against sequences in the nonredundant Swiss Protein Database. HtxB to HtxN are each homologous to the corresponding PhnC to PhnP components of the E. coli C—P lyase, except that homologs to PhnO and PhnF, which are not required for phosphonate utilization in E. coli (21), are absent (Table 3). Thus, all of the genes required to encode a functional C—P lyase are present in a 10.9-kbp region immediately downstream of HtxA. The htx operon not only is homologous to the E. coli phn operon at the level of the predicted amino acid sequences but also conserves the order of the E. coli C—P lyase genes (minus those that are absent). The downstream orf282 and orf344 genes do not encode Phn-like proteins but instead are homologous to a hypothetical transmembrane protein and a putative cointegrate resolution protein, respectively (Table 3).
TABLE 3.
Comparison of the Htx and Phn proteins in P. stutzeri with each other and with the Phn proteins of E. coli
| Phn protein | Closest homolog of predicted product (% identity)a | % Identity to E. coli Phn homologb | Htx protein | Closest homolog of predicted product(s) (% identity) | % Identity to E. coli Phn homologb | Phn and Htx homologs of P. stutzeri WM88c | % Identity between Phn and Htx proteins | Proposed function of Phn and Htx proteins |
|---|---|---|---|---|---|---|---|---|
| PhnD | P. aeruginosa (PAO1) PhnD (73.7) | 54.0 | HtxB | Nostoc sp. PhnD, phosphonate binding protein (25.7) | 18.1 | PhnD/HtxB | 20.3 | Phosphonate binding protein of ABC transporter |
| PhnE | P. aeruginosa (PAO1) PhnE (85.9) | 69.2 | HtxC | Bradyrhizobium japonicum PhnE (40.1) | 32.6 | PhnE/HtxC | 33.3 | Phosphonate transport permease |
| HtxE | Bacillus anthracis PhnE (41.3) | 26.8 | PhnE/HtxE | 29.8 | ||||
| PhnC | P. syringae (pv. tomato) PhnC (80.8) | 58.3 | HtxD | E. coli PhnC (44.7) | 44.7 | PhnC/HtxD | 20.3 | Phosphonate binding protein of ABC transporter |
| PhnF | P. aeruginosa (PAO1) probable transcriptional regulator (67.9) | 41.1 | —d | Transcriptional regulator (GntR family) | ||||
| PhnG | P. syringae (pv. tomato) PhnG (73.2) | 43.9 | HtxF | Sinorhizobium sp. PhnG (33.3) | 21.4 | PhnG/HtxF | 22.6 | Phosphonate metabolism protein |
| PhnH | P. syringae (pv. tomato) PhnH (61.0) | 33.2 | HtxG | E. coli PhnH (32.6) | 32.6 | PhnH/HtxG | 25.4 | Phosphonate metabolism protein |
| PhnI | P. syringae (pv. tomato) PhnI (80.8) | 62.9 | HtxH | Mesorhizobium loti PhnI (45.2) | 43.6 | PhnI/HtxH | 42.6 | Phosphonate metabolism protein |
| PhnJ | P. syringae (pv. tomato) PhnJ (89.5) | 75.4 | HtxI | E. coli PhnJ (53.7) | 53.7 | PhnJ/HtxI | 53.0 | Phosphonate metabolism protein |
| PhnK | P. aeruginosa (PAO1) putative ATP binding component (86.3) | 65.2 | HtxJ | Agrobacterium tumefaciens PhnK (40.2) | 32.3 | PhnK/HtxJ | 30.5 | Phosphonate ATP binding protein |
| PhnL | P. aeruginosa (PAO1) putative ATP binding component (86.0) | 58.5 | HtxK | M. loti PhnL (52.0) | 42.6 | PhnL/HtxK | 42.6 | Phosphonate ATP binding protein |
| PhnM | P. syringae (pv. tomato) PhnM (71.6) | 56.5 | HtxL | Rhizobium luguminosarum PhnM (36.6) | 34.7 | PhnM/HtxL | 35.0 | Phosphonate metabolism protein |
| PhnN | P. syringae (pv. tomato) PhnN (62.0) | 39.4 | HtxM | S. meliloti PhnN (47.3) | 41.6 | PhnN/HtxM | 37.4 | Phosphonate metabolism accessory protein |
| PhnP | P. syringae (pv. tomato) PhnP (71.5) | 46.0 | HtxN | P. aeruginosa conserved hypothetical protein (53.0); P. syringae PhnP (51.0) | 45.9 | PhnP/HtxN | 49.0 | Phosphonate metabolism accessory protein |
Closest homologs were determined with FASTA searches of the predicted amino acid sequences against sequences in the nonredundant Swiss Protein Database on 15 May 2003.
Pairwise sequence alignments were performed with the predicted amino acid sequences of the P. stutzeri Phn or Htx protein with the E. coli Phn homologs.
Pairwise sequence alignments were performed with the predicted amino acid sequences of the P. stutzeri Phn or Htx proteins.
The Phn homolog was absent from the htx sequence.
The htxA gene and the putative C—P lyase-encoding genes comprise a single transcriptional unit.
The close proximity and/or overlap of the ORFs within the htx locus suggest that they form a single transcriptional unit. Of the 14 ORFs identified, 5 have coding regions that overlap, whereas the remaining ORFs are separated by, at most, 44 nucleotides, with the exception of htxBC, which is separated by 125 nucleotides.
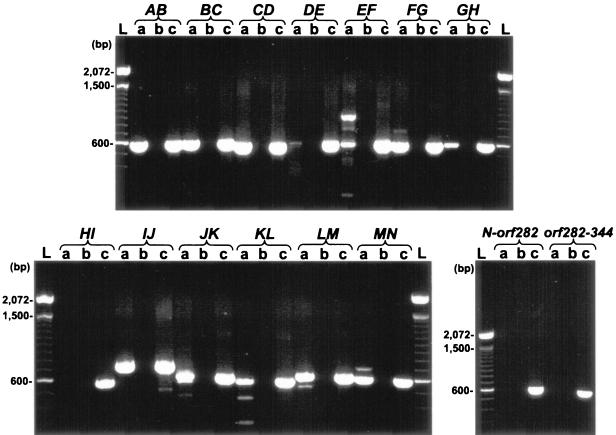
To test whether these genes are cotranscribed, RT-PCR was used to determine the presence of the junction sequences between each gene in total RNA isolated from hypophosphite-grown P. stutzeri (Fig. 2). Amplification products were seen for the intergenic regions between all adjacent htx genes, with the exception of htxHI. Nevertheless, because htxH and htxI overlap by 5 bp, it is likely that they are cotranscribed. Therefore, the htxABCDEFGHIJKLMN genes almost certainly comprise an operon. Amplification products were not observed for the regions between the htxN, orf282, and orf344 genes, which, in conjunction with their lack of homology to the known P assimilation genes, suggests that they are not within the same transcriptional unit as the htx genes.
FIG. 2.
RT-PCR of total RNA prepared from P. stutzeri WM567 grown on hypophosphite as the sole P source to determine the operon structure of htx. Lanes a show complete RT reactions; lanes b contain a negative control, with which no reverse transcriptase was added to the reaction mixture; and lanes c contain a PCR-positive control, for which chromosomal DNA was used as the template. Lanes L, 100-bp ladder. The junction sequences amplified are indicated above each set of reactions. For a list of the primers used and the predicted PCR product sizes, refer to Table 2.
Identification of a second C—P lyase-encoding locus in P. stutzeri.
To investigate whether htxB to htxN encode a functional C—P lyase, and to determine the role of this putative C—P lyase with respect to phosphite and phosphonate metabolism, a strain with a deletion of the entire region encompassing the htx and ptx operons (ΔptxA-orf344 mutant) was constructed. Consistent with previous results (22), the deletion mutant was no longer able to grow on hypophosphite; however, growth on methylphosphonate and aminoethylphosphonate was still observed. Furthermore, after prolonged incubation, a very low level of growth was observed on phosphite as the sole P source. These data suggest the presence of an additional pathway(s) for the utilization of phosphonates and phosphite.
The genes permitting growth on phosphonates and phosphite in the ΔptxA-orf344 strain were identified by in vivo Tn5-RL27 transposon mutagenesis as described previously (14). A Tn5 insertion mutant unable to grow on methylphosphonate or phosphite as the sole P source was isolated, and the sequence with mutation was cloned. The sequence adjacent to the Tn5-RL27 insertion showed that the transposon was inserted into an E. coli phnI homolog. Additional sequencing of the flanking DNA revealed the presence of 13 ORFs that also share significant predicted amino acid sequence identity with putative Phn proteins of Pseudomonas aeruginosa and Pseudomonas syringae (Table 3). Both the sequence and the organization of the 13 ORFs are homologous to the genetically characterized phn operon of E. coli, except that phnO is absent from the P. stutzeri phn sequence. The inability of the ΔptxA-orf344 phnI::Tn5-RL27 mutant to utilize methylphosphonate as a sole P source, in contrast to its parent, indicates that the P. stutzeri phnCDEFGHIJKLMNP operon encodes a functional C—P lyase.
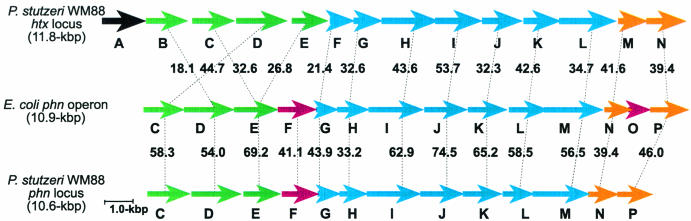
Interestingly, the genes constituting this operon are distinctly different from the phn-like genes htxB to htxN. Pairs of predicted amino acid sequences were aligned to compare the P. stutzeri htx-encoded C—P lyase components with the corresponding P. stutzeri phn-encoded C—P lyase subunits (Table 3). The P. stutzeri Phn proteins share significant identity with the Phn proteins of other pseudomonads (61 to 90% identity) and with E. coli (39 to 75%) but share much less identity with the htx-encoded Phn-like proteins (20 to 53%), which are generally more similar to the Phn components of bacteria from the family Rhizobiaceae. This trend is also true with respect to the organization of the genes in the htx and phn operons of P. stutzeri in comparison to that of E. coli (Fig. 3). The P. stutzeri phn operon is quite similar to that of E. coli, missing only a homolog to E. coli phnO. In contrast, the htx operon not only contains an additional phnE homolog, but also is missing homologs to the putative regulators encoded by both phnF and phnO.
FIG. 3.
Organization of the genes of the htx and phn operons involved in the metabolism of phosphonates in P. stutzeri compared to that of the phn operon of E. coli. The black arrow represents genes within the htx operon that are not homologous to any phn gene, green arrows represent genes likely involved in phosphonate transport, red arrows represent genes with putative regulatory function, blue arrows represent genes thought to encode the catalytic components of the C—P lyase, and gold arrows represent genes believed to encode accessory proteins to C—P lyase. The percentages of predicted amino acid sequence identity between each of the homologous proteins are indicated.
The roles of the htx and phn operons in the oxidation of reduced P compounds.
The roles of the phnC to -P and htxA to -N operons in the metabolism of reduced P compounds were further examined using a series of mutants harboring complete deletions of phnC to phnP (Δphn), ptxA to ptxE (Δptx), and/or htxA to orf344 (Δhtx) as single, double, or triple mutants. Each mutant was tested for its ability to utilize a variety of different reduced P compounds as sole P sources (Table 4).
TABLE 4.
Growth of P. stutzeri Δhtx, Δptx, and Δphn mutants on various P sources
| Phosphorus source | Growtha
|
|||||||
|---|---|---|---|---|---|---|---|---|
| WM567 (WT) | WM1926 (Δhtx) | WM3746 (Δptx) | WM3614 (Δphn) | WM3747 (Δhtx Δptx) | WM3748 (Δphn Δptx) | WM3616 (Δhtx Δphn) | WM3617 (δptx-htx Δphn) | |
| Phosphate | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| Phosphite | +++ | +++ | +b | ++ | +/−b | − | +++ | − |
| Hypophosphite | +++ | +/− | +b | +++ | +/−b | − | +/− | − |
| Methylphosphonate | ++ | +++ | ++ | + | +++ | + | − | − |
| Aminoethylphosphonate | ++ | +++ | ++ | − | +++ | − | − | − |
| Phenylphosphonate | − | − | − | − | − | − | − | − |
| Phenylphosphinate | − | − | − | − | − | − | − | − |
| Dimethylphosphinate | − | − | − | − | − | − | − | − |
| Glyphosate | − | − | − | − | − | − | − | − |
| Phosphomycin | − | − | − | − | − | − | − | − |
+++, growth equal to that of the positive-control strain WM567 on 0.5 mM K2HPO4 after 2 days of incubation at 37°C; ++, growth equal to that of the positive control after 4 days of incubation at 37°C; +, growth equal to that of the positive control after 7 days of incubation at 37°C; +/−, growth exceeding that of the negative control but not equal to that of the positive control after 7 days of incubation at 37°C; −, growth equal to that of the negative-control strain WM567 on phosphate-free agar without an added P source after 7 days of incubation at 37°C. WT, wild type.
Mutant colonies grew above the background growth for this sample.
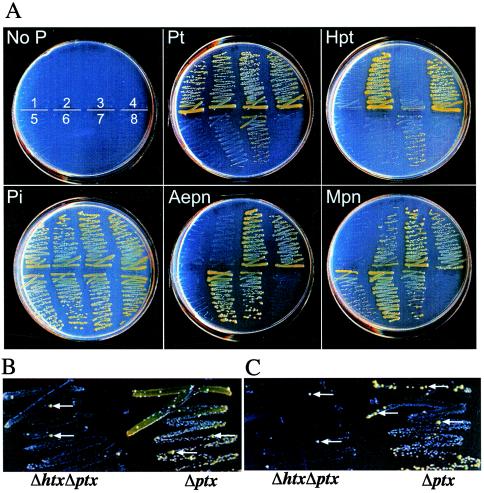
Growth of the Δphn Δptx strain on methylphosphonate as the sole source of P indicates that the putative C—P lyase system encoded by the htx operon is functional. However, the growth conferred by the htx-encoded C—P lyase is poor compared to that of the Δhtx Δptx strain, which relies on the phn-encoded C—P lyase system for methylphosphonate utilization (Fig. 4, compare streaks 5 and 6). Also, the htx-encoded system confers growth on methylphosphonate but does not allow growth on aminoethylphosphonate, whereas the phn-encoded C—P lyase confers growth on both substrates. The substrate specificities of both P. stutzeri C—P lyase systems are rather narrow relative to those of other known C—P lyase systems (26, 31); no growth was observed by any of the strains on phenylphosphonate, phenylphosphinate, dimethylphosphinate, glyphosate, or phosphonomycin, even after an incubation time of greater than 10 days. These differences in substrate utilization may reflect the specificity of the C—P lyase enzyme and/or the associated phosphonate transport systems or, alternatively, may reflect differences in expression of the two operons in response to different substrates. Deletion of both htx and phn abolishes growth on all phosphonates, indicating that these are the only two pathways present in P. stutzeri for the utilization of phosphonates.
FIG. 4.
Growth of P. stutzeri htx, ptx, and phn deletion mutants on minimal media with various P sources after 4 days of growth. (A) Fresh cultures were streaked from MOPS minimal solid agar containing 0.1 mM Pi onto MOPS minimal agar containing the indicated P sources (phosphite [Pt], hypophosphite [Hpt], inorganic phosphate [Pi], aminoethylphosphonate [Aepn], and methylphosphonate [Mpn]) at 0.5 mM. The schematic on the plate lacking phosphorus (No P) represents the order in which the deletion mutants were streaked: 1, Δhtx Δphn mutant (WM3616); 2, Δphn mutant (WM3614); 3, Δhtx mutant (WM1926); 4, wild type (WM567); 5, Δphn Δptx mutant (WM3748); 6, Δhtx Δptx mutant (WM3747); 7, Δptx mutant (WM3746); and 8, Δptx-htx Δphn mutant (WM3617) (B and C). Representative Δhtx Δptx and Δptx mutant colonies showing enhanced growth that were grown on phosphite (B) and hypophosphite (C) are indicated by white arrows.
As previously observed, strains with the ptx genes deleted retain a small amount of growth on phosphite after 7 days of incubation. This growth is absent in the Δptx Δphn mutant, consistent with a role for phn in phosphite oxidation, as has been previously described for E. coli (20). Interestingly, the lack of growth on phosphite for the Δptx Δphn mutant also demonstrates that the htx-encoded C—P lyase is incapable of phosphite oxidation.
Strains that rely on phn for phosphite oxidation give rise to better-growing mutants during growth with phosphite or hypophosphite as the sole P source (Fig. 4B and C). The responsible mutations are most likely within the phn operon, because such mutations were not observed in Δphn strains. Interestingly, the absence of similar mutations in the Δphn Δptx mutant suggests that the htx operon cannot be mutated to allow phosphite oxidation. Strains in which ptx remains intact also do not give rise to better-growing mutants; however, ptx-dependent growth on phosphite is much more robust than phn-dependent growth.
Surprisingly, it appears that the htx-encoded C—P lyase interferes with phn-encoded phosphonate utilization. Thus, strains from which the htx operon is deleted grow faster on phosphonates than does the wild type (Fig. 4, streak 3 or 6 compared to streak 4). A similar effect is not observed in Δphn strains, indicating that the phn-encoded C—P lyase does not negatively impact the htx-encoded C—P lyase. An even more pronounced effect is observed when plasmids with the P. stutzeri htx operon are carried in E. coli. In this case, phn+ E. coli strains become completely incapable of phosphonate utilization (data not shown). This negative interaction is specific for growth on phosphonates and is not observed during growth on phosphite. Instead, the opposite was observed; i.e., growth on phosphite was better in the presence of the htx operon than in its absence (compare streak 7 to streak 6), despite the observation that the presence of htx alone did not allow the strain to grow on phosphite. This result suggests that some subset of genes with the htx operon can function in concert with phn operon products to promote phosphite oxidation.
DISCUSSION
The data presented here show that P. stutzeri encodes two discrete C—P lyases that allow the use of phosphonates as sole P sources. Although the presence of multiple C—P lyases within the same host has previously been suggested based on the assimilation kinetics of different phosphonic acid substrates (12), this is the first case in which multiple operons have been definitively identified and phenotypically characterized. Interestingly, there are significant differences between the htx-encoded and phn-encoded C—P lyases. The proteins encoded by the phn and htx operons are more similar to homologs from other bacteria than they are to each other, with the phn gene products being most similar to homologs from other pseudomonads (62 to 90%) and the htx operon being more similar to homologs found in members of the Rhizobiaceae (33 to 52%). These data strongly suggest that the two operons evolved in different hosts, rather than by duplication of the same operon within the chromosome. Further, given the homology of the phn operon to those of other pseudomonads, it seems likely that the htx operon was the more recent addition to the P. stutzeri genome.
Additional differences were observed in the substrate specificities of the two C—P lyase systems. The phn-encoded C—P lyase system allows growth on methylphosphonate, aminoethylphosphonate, and phosphite, while the htx-encoded C—P lyase system allows growth only on methylphosphonate. Both systems have relatively narrow substrate profiles relative to those of other known C—P lyase systems: neither operon allowed growth on phenylphosphonate, glyphosate, or various phosphinates, although these substrates are widely used by other bacteria (26, 31). It is important to note that this substrate specificity is not necessarily a property of the C—P lyase enzyme. Differences in substrate specificity of the associated phosphonate transporters could also account for these data, as could differences in the expression of the two operons in response to different substrates.
Further evidence that phn and htx evolved in different organisms stems from the observation that the htx operon has a negative effect on the function of the phn-encoded C—P lyase. This negative interaction appears to be one-sided, in that deletion of htx improves growth on phosphonates conferred by phn but deletion of phn does not improve growth on methylphosphonate conferred by htx. Further, the negative effect of htx is not specific for the P. stutzeri phn operon and is even more pronounced in E. coli. Although the nature of this negative interaction is unknown, there are several possible explanations for this phenotype. It has been postulated that C—P lyase is a membrane-associated enzyme complex. This supposition is supported by the observation that C—P lyase activity in whole-cell suspensions does not require phnCDE for phosphonate transport (35). Given the likelihood that the components of C—P lyase interact in some way to form an active complex, it seems possible that the respective Htx and Phn proteins are similar enough to each other to form hybrid, inactive complexes. Such negative interactions may also exist in the assembly of the binding protein-dependent transporters encoded by htxBCDE and phnCDE. The existence of dominant negative mutants incapable of assembling the homologous E. coli maltose transport system suggests that such a scenario is possible (24). Similar interactions between htxBCDE and phnCDE might result in the formation of a hybrid, inactive transport system, resulting in deficient phosphonate uptake. Careful analysis of the nature of this dominant negative effect may significantly add to our understanding of the C—P lyase reaction, revealing how the many proteins required for catalysis and/or transport interact with one another.
We had anticipated that both the htx- and phn-encoded C—P lyases would allow phosphite oxidation, as does the C—P lyase of E. coli; however, our data indicate that only the phn-encoded C—P lyase has this property. Despite this ability, we conclude that phosphite oxidation is predominantly a function of the ptx operon, in agreement with the results of a previous study (22). Thus, phn-dependent growth on phosphite is much poorer than ptx-dependent growth. Further, ptx deletion strains, which depend on the phn operon for phosphite oxidation, give rise to mutants with better growth on hypophosphite or phosphite (Fig. 4B and C). This finding suggests that the phn operon has not been subject to selective pressure for better phosphite utilization since the acquisition of the ptx operon. In this context, it should be noted that, while most pseudomonads possess a phn operon (based on available genomic sequences), the htx and ptx loci appear to be uncommon and are more likely to be recent acquisitions. Thus, although pseudomonads with these genes are easily isolated by selective enrichment, none of the published genomes contain these genes, nor do six authentic P. stutzeri strains from the American Type Culture Collection (22).
The observation that the htx operon does not allow growth on phosphite contradicts our hypothesis that this operon encodes a complete pathway for hypophosphite oxidation. Nevertheless, our data do suggest a potential linkage between the use of hypophosphite and phosphonates, because the htx-encoded C—P lyase is cotranscribed with the hypophosphite-oxidizing enzyme encoded by htxA. Three explanations for this linkage seem plausible. First, our original hypothesis may have been correct at an earlier stage in evolution. Accordingly, it is possible that the htx operon once provided phosphite oxidation activity and that over time, more-efficient pathways for the oxidation of phosphite, such as ptx and phn, were introduced in P. stutzeri. Alternatively, a phosphite-oxidizing htx operon may have been introduced into a strain that already possessed phn and/or ptx. In either case, this introduction may have resulted in the loss of the phosphite oxidation phenotype conferred by htx due to the lack of selective pressure to maintain it. Moreover, the negative interaction between the two C—P lyases may have provided additional pressure for loss-of-function mutations in the htx operon. A second explanation suggests that the htx operon encodes a two-step pathway for the oxidation-reduced P compounds having both carbon—phosphorus and hydrogen—phosphorus bonds, such as (C—P—H) phosphinates (P valence, +1). One such compound, demethylphosphinothricin, is known to be produced by several Streptomyces species during the synthesis of the antibiotic bialaphos and is likely present in the soil (25). Such a pathway would involve the oxidation of the C—P—H bonds by HtxA to produce a phosphonate (C—P—OH), which would then be oxidized to phosphate by the htx-encoded C—P lyase. Unfortunately, our attempts to test this idea by examining growth on two commercially available phosphinates, dimethylphosphinate and phenylphosphinate, failed due to the lack of growth on either substrate. Nevertheless, as-yet-unidentified phosphinate substrates for this putative HtxA/C—P lyase pathway may exist. Finally, it may be that HtxA and C—P lyase are cotranscribed only because both pathways are used to acquire P from less favorable sources and, thus, are regulated by a common phosphate starvation-inducible promoter. Their close proximity may simply be due to a grouping of related genes whose expression is under the control of the same regulators.
Acknowledgments
We thank Marlena Wilson and Jill Bradshaw for assistance in strain construction.
This work was supported by grant GM59334 from the National Institute of General Medical Sciences.
REFERENCES
- 1.Adams, F., and J. P. Conrad. 1953. Transition of phosphite to phosphate in soils. Soil Sci. 75:361-371. [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1992. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 3.Casida, L. E., Jr. 1960. Microbial oxidation and utilization of orthophosphite during growth. J. Bacteriol. 80:237-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, C. M., Q. Z. Ye, Z. M. Zhu, B. L. Wanner, and C. T. Walsh. 1990. Molecular biology of carbon-phosphorus bond cleavage. Cloning and sequencing of the phn (psiD) genes involved in alkylphosphonate uptake and C—P lyase activity in Escherichia coli B. J. Biol. Chem. 265:4461-4471. [PubMed] [Google Scholar]
- 5.Clark, L. L., E. D. Ingall, and R. Benner. 1999. Marine organic phosphorus cycling: novel insights from nuclear magnetic resonance. Am. J. Sci. 299:724-737. [Google Scholar]
- 6.Costas, A. M., A. K. White, and W. W. Metcalf. 2001. Purification and characterization of a novel phosphorus-oxidizing enzyme from Pseudomonas stutzeri WM88. J. Biol. Chem. 276:17429-17436. [DOI] [PubMed] [Google Scholar]
- 7.Foster, T. L., and L. Winans, Jr. 1977. Anaerobic utilization of phosphite/phosphine as a sole source of phosphorus: implication to growth in the Jovian environment, p. 81-86. In R. Holmquist and A. C. Stickland (ed.), Life sciences and space research, XV: proceedings of the Open Meeting of the Working Group on dbchlochSpace Biology of the Nineteenth Plenary Meeting of COSPAR, Philadelphia, Pa., 1976. Pergamon Press, New York, N.Y. [PubMed]
- 8.Foster, T. L., L. Winans, Jr., and S. J. S. Helms. 1978. Anaerobic utilization of phosphite and hypophosphite by Bacillus sp. Appl. Environ. Microbiol. 35:937-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heinen, W., and A. M. Lauwers. 1974. Hypophosphite oxidase from Bacillus caldolyticus. Arch. Microbiol. 95:267-274. [Google Scholar]
- 10.Horiguchi, M. 1984. Occurrence, identification and properties of phosphonic and phosphinic acids, p. 24-52. In T. Hori, M. Horiguchi, and A. Hayashi (ed.), Biochemistry of natural C—P compounds. Japanese Association for Research on the Biochemistry of C—P Compounds, Shiga, Japan.
- 11.Hove-Jensen, B., T. J. Rosenkrantz, A. Haldimann, and B. L. Wanner. 2003. Escherichia coli phnN, encoding ribose 1,5-bisphosphokinase activity (phosphoribosyl diphosphate forming): dual role in phosphonate degradation and NAD biosynthesis pathways. J. Bacteriol. 185:2793-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kertesz, M., A. Elgorriaga, and N. Amrhein. 1991. Evidence for two distinct phosphonate-degrading enzymes (C—P lyases) in Arthrobacter sp. GLP-1. Biodegradation 2:53-59. [DOI] [PubMed] [Google Scholar]
- 13.Kolowith, L. C., E. D. Ingall, and R. Benner. 2001. Composition and cycling of marine organic phosphorus. Limnol. Oceanogr. 46:309-320. [Google Scholar]
- 14.Larsen, R. A., M. M. Wilson, A. M. Guss, and W. W. Metcalf. 2002. Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Arch. Microbiol. 178:193-201. [DOI] [PubMed] [Google Scholar]
- 15.Malacinski, G., and W. A. Konetzka. 1966. Bacterial oxidation of orthophosphite. J. Bacteriol. 91:578-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mastalerz, P., Z. Wieczorek, and M. Kochman. 1965. Utilization of carbon-bound phosphorus by microorganisms. Acta Biochim. Pol. 12:151-156. [PubMed] [Google Scholar]
- 17.McMullan, G., R. Watkins, D. B. Harper, and J. P. Quinn. 1991. Carbon-phosphorus bond cleavage activity in cell-free extracts of Enterobacter aerogenes ATCC 15038 and Pseudomonas sp. 4ASW. Biochem. Int. 25:271-279. [PubMed] [Google Scholar]
- 18.Metcalf, W. W., W. Jiang, L. L. Daniels, S. K. Kim, A. Haldimann, and B. L. Wanner. 1996. Conditionally replicative and conjugative plasmids carrying lacZ alpha for cloning, mutagenesis, and allele replacement in bacteria. Plasmid 35:1-13. [DOI] [PubMed] [Google Scholar]
- 19.Metcalf, W. W., and B. L. Wanner. 1993. Evidence for a fourteen-gene, phnC to phnP locus for phosphonate metabolism in Escherichia coli. Gene 129:27-32. [DOI] [PubMed] [Google Scholar]
- 20.Metcalf, W. W., and B. L. Wanner. 1991. Involvement of the Escherichia coli phn (psiD) gene cluster in assimilation of phosphorus in the form of phosphonates, phosphite, Pi esters, and Pi. J. Bacteriol. 173:587-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Metcalf, W. W., and B. L. Wanner. 1993. Mutational analysis of an Escherichia coli fourteen-gene operon for phosphonate degradation, using TnphoA′ elements. J. Bacteriol. 175:3430-3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Metcalf, W. W., and R. S. Wolfe. 1998. Molecular genetic analysis of phosphite and hypophosphite oxidation by Pseudomonas stutzeri WM88. J. Bacteriol. 180:5547-5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mourez, M., M. Hofnung, and E. Dassa. 1997. Subunit interactions in ABC transporters: a conserved sequence in hydrophobic membrane proteins of periplasmic permeases defines an important site of interaction with the ATPase subunits. EMBO J. 16:3066-3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murakami, T., H. Anzai, S. Imai, A. Satoh, K. Nagaoka, and C. J. Thompson. 1986. The bialaphos biosynthetic genes of Streptomyces hygroscopicus: molecular cloning and characterization of the gene cluster. Mol. Gen. Genet. 205:42-50. [Google Scholar]
- 26.Parker, G. F., T. P. Higgins, T. Hawkes, and R. L. Robson. 1999. Rhizobium (Sinorhizobium) meliloti phn genes: characterization and identification of their protein products. J. Bacteriol. 181:389-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schink, B., and M. Friedrich. 2000. Phosphite oxidation by sulphate reduction. Nature 406:37. [DOI] [PubMed] [Google Scholar]
- 29.Seto, H., and T. Kuzuyama. 1999. Bioactive natural products with carbon-phosphorus bonds and their biosynthesis. Nat. Prod. Rep. 16:589-596. [DOI] [PubMed] [Google Scholar]
- 30.Ternan, N. G., J. W. McGrath, G. McMullan, and J. P Quinn. 1998. Organophosphonates: occurrence, synthesis and biodegradation by microorganisms. World J. Microbiol. Biotechnol. 14:635-647. [Google Scholar]
- 31.Wackett, L. P., S. L. Shames, C. P. Venditti, and C. T. Walsh. 1987. Bacterial carbon-phosphorus lyase: products, rates, and regulation of phosphonic and phosphinic acid metabolism. J. Bacteriol. 169:710-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wanner, B. L. 1986. Novel regulatory mutants of the phosphate regulon in Escherichia coli K-12. J. Mol. Biol. 191:39-58. [DOI] [PubMed] [Google Scholar]
- 33.Wanner, B. L., and J. A. Boline. 1990. Mapping and molecular cloning of the phn (psiD) locus for phosphonate utilization in Escherichia coli. J. Bacteriol. 172:1186-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.White, A. K., and W. W. Metcalf. 2002. Isolation and biochemical characterization of hypophosphite/2-oxoglutarate dioxygenase. A novel phosphorus-oxidizing enzyme from Pseudomonas stutzeri WM88. J. Biol. Chem. 277:38262-38271. [DOI] [PubMed] [Google Scholar]
- 35.Yakovleva, G. M., S. K. Kim, and B. L. Wanner. 1998. Phosphate-independent expression of the carbon-phosphorus lyase activity of Escherichia coli. Appl. Microbiol. Biotechnol. 49:573-578. [DOI] [PubMed] [Google Scholar]