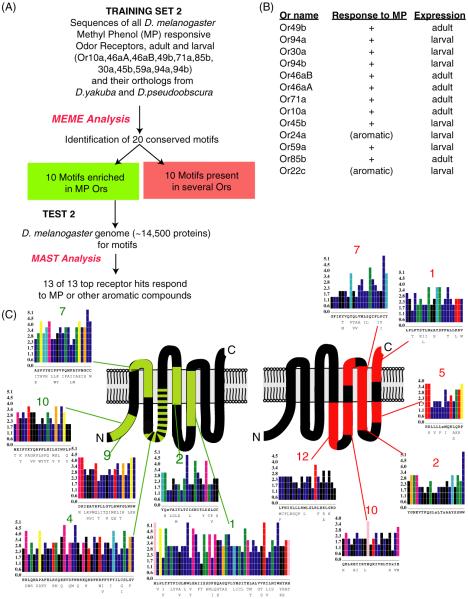
Figure 2.
Computational identification of motifs that are phenol-specific and family-wide. (A) Schematic of a computational approach to identify amino acid motifs shared among odor receptors that respond to phenols 2-methylphenol or 4-methylphenol indicated as MP. (B) Odor receptors that are identified from scanning the Drosophila genome using conserved amino acid motifs enriched in phenol odor receptors. (C) Schematic representing the locations of the phenol-specific motifs and the family-wide motifs on a secondary structure model of odor receptors. The most common amino acid compositions are indicted below each position on the X-axis and their MEME-determined conservation score indicated on Y-axis for the best-conserved sequence motifs.