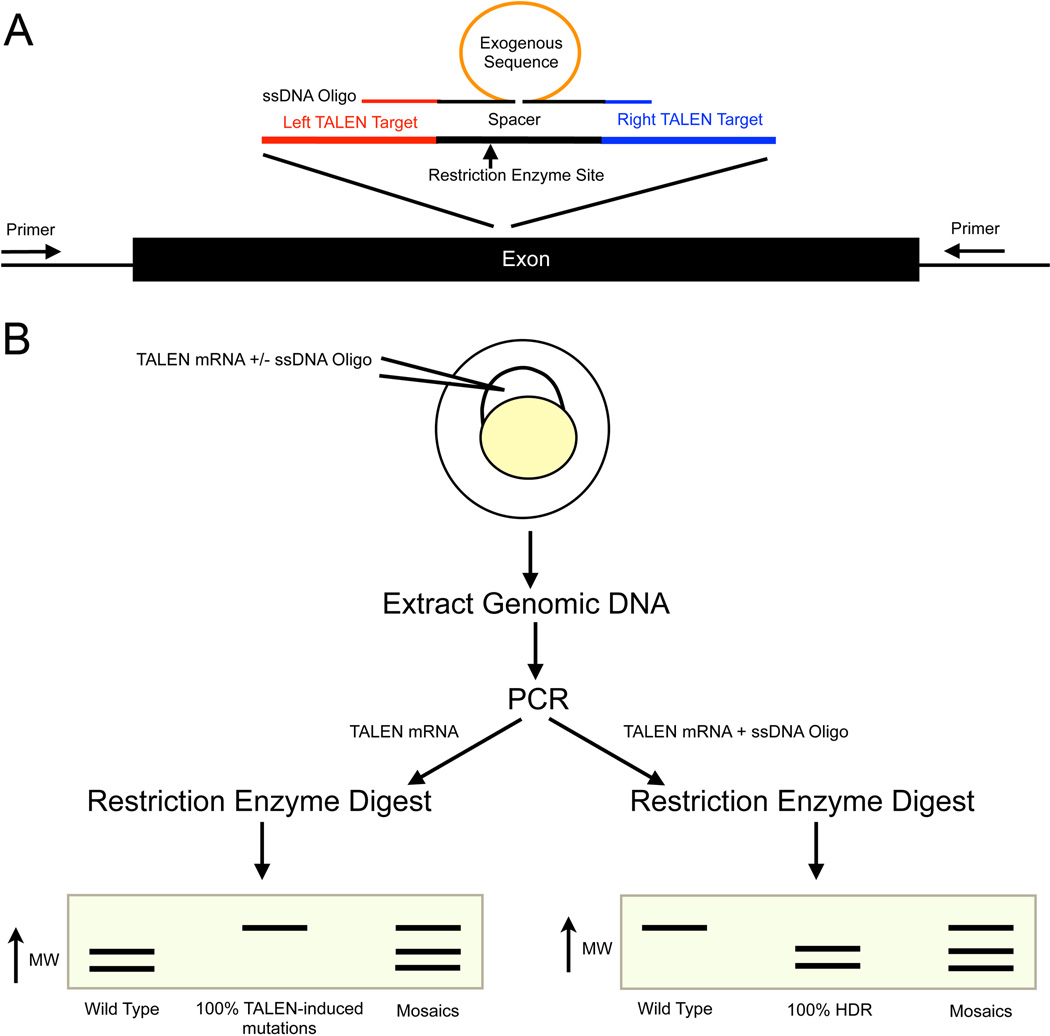
Figure 2. Experimental design of zebrafish knockin and knockout experiments.
A) A diagram of a genetic locus to be targeted. To create a knockout mutation, TALENs should target a conserved exon early exon. Exon one is often an isoform-specific exon, so it is often better to target a downstream exon. Two TALENs (left and right) bind to the genomic sequence on opposite strands and create a double-strand break. Within the spacer region, we typically design around a local unique restriction enzyme for simplified downstream screening. For knockin experiments, a ssDNA oligo is designed as shown. The homology arms do not normally include the entire TALEN binding site. An exogenous sequence can be added to the center of the spacer region. B) Experimental design for creating knockout or knockin zebrafish. Inject TALEN mRNA to create a knockout or inject TALEN mRNA plus ssDNA oligo for HDR. Extract the genomic DNA from the embryos, perform a PCR and subsequently digest the PCR product. For knockout experiments (injected only with TALEN mRNA), a mutation will result in loss of the restriction enzyme binding site and a larger band on the enzyme analysis on the PCR product. For the knockin experiments such as when adding a new restriction enzyme site to the spacer region, successful HDR results in a digested band and unmodified (wild type) is therefore uncut in the enzymatic assay.