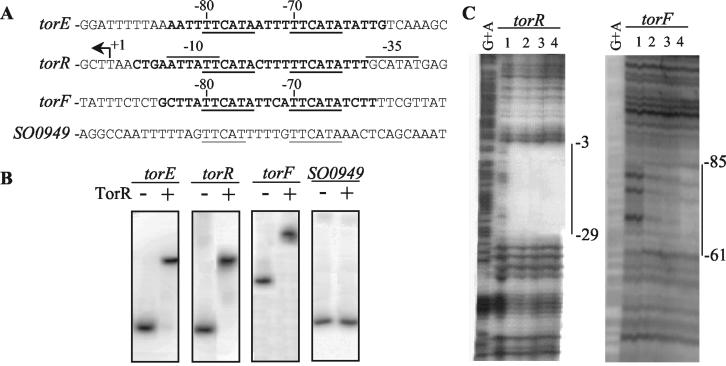
FIG. 2.
(A) Alignment of the torE, torR, torF, and SO0949 promoter regions. The regions protected by TorR are indicated in bold. The direct repeats are underlined. Positions relative to the transcription start sites are indicated above the sequences. For convenience, the complementary sequence of the torR promoter is presented. The direct repeat sequence of SO0949 is centered at −165 bp from the initiation codon. (B) Electrophoretic gel shift analysis of TorR interaction with the torE, torR, torF, and SO0949 promoters. The DNA fragments containing the torE (position −90 to + 119 relative to the transcription start site), torR (position −159 to + 42 relative to the transcription start site), torF (position −306 to + 80 relative to the transcription start site), and SO0949 (position −234 to −54 relative to the initiation codon) promoter regions were obtained by PCR with the primer pairs Wt-Erev, pR2-pR1, F3-F5, and 949A-949B, respectively. The labeled fragments were used in gel shift experiments in the presence (+) or absence (−) of a 1 μM concentration of purified TorR protein. (C) Analysis of TorR binding to the torR and the torF promoter regions by DNase I footprinting experiments. The DNA fragments corresponding to the torR and the torF promoter regions were obtained by PCR by using the primer pairs labeled pR2-unlabeled pR1 and labeled F5-unlabeled F3, respectively. The labeled DNA fragments were digested with DNase I in the presence of the following concentrations of TorR protein: lane 1, no protein; lane 2, 0.25 μM; lane 3, 1 μM; and lane 4, 2.5 μM. The G+A sequencing ladders are shown, and the vertical bars indicate the protected regions.