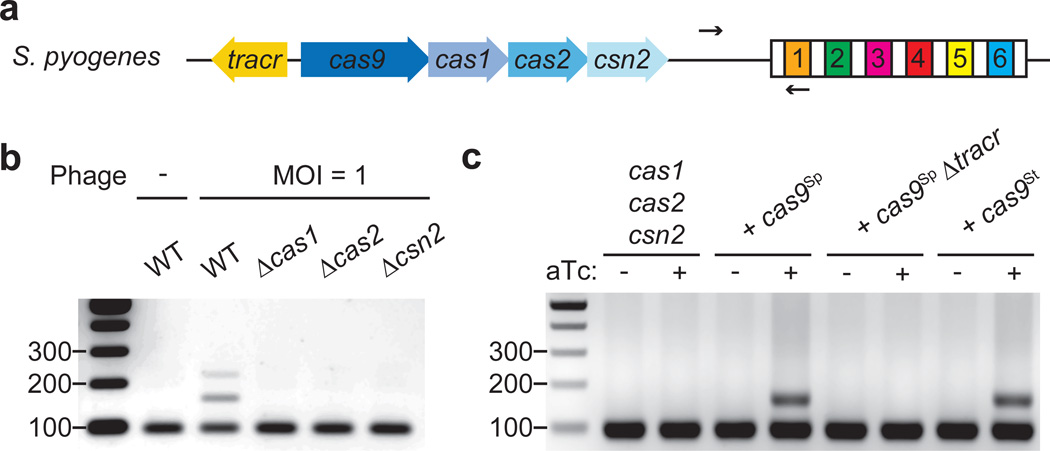
Figure 1. Cas9 is required for spacer acquisition.
a, Organization of the S. pyogenes type II CRISPR-Cas locus. Arrows indicate the annealing position of the primers used to check for the expansion of the CRISPR array. b, PCR-based analysis of liquid cultures to check for the acquisition of new spacer sequences in the presence or the absence of phage ϕNM4γ4 infection. Wild-type (WT) as well as different cas mutants were analyzed. Image is representative of three technical replicates. MOI; multiplicity of infection. c, Cultures over-expressing Cas1, Cas2 and Csn2 under the control of a tetracycline-inducible promoter were analyzed using PCR for spacer acquisition in the absence of phage infection. The strain was complemented with plasmids carrying either St or Sp Cas9 (see Extended Data Fig. 3), in the last case with or without the tracrRNA gene (Δtracr). Image is representative of three technical replicates. aTc; anhydrotetracycline.