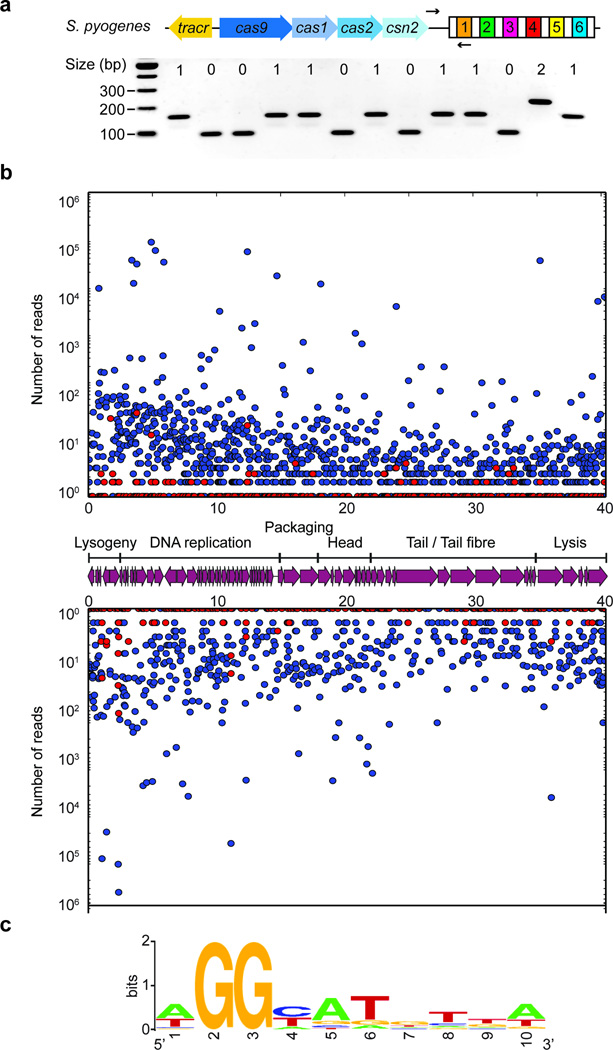
Extended Data Figure 1. The S. pyogenes type II CRISPR-Cas system displays a strong bias for the acquisition of spacers matching viral protospacers with NGG PAMs.
a, Analysis of bacteriophage-insensitive mutant colonies using PCR and agarose gel electrophoresis, representative of five technical replicates. Bacteria and phage were mixed in top agar and incubated overnight. DNA was isolated from individual colonies resistant to phage infection and used as template for a PCR reaction with primers (arrows) H182 and H183 (Extended Data Table 2), which amplify the 5’ end of the S. pyogenes CRISPR array. The size of the PCR band indicates the number of new spacers (shown at the top of the gel). Cells without additional spacers resist infection by a CRISPR-independent mechanisms, presumably envelope resistance. b, Analysis of acquired spacers during phage infection of a population of bacteria carrying the S. pyogenes type II CRISPR-Cas system. Liquid cultures of bacteria were infected with phage, surviving cells were collected at the end of the infection, DNA extracted and used as template for a PCR reaction as described above. Amplification products were separated by agarose gel electrophoresis and the DNA of the bands corresponding to products with additional spacers was extracted and sent for Mi-Seq next generation sequencing. Reads corresponding to newly acquired spacers were plotted according to their position in the phage ϕNM4γ4 genome (x-axis) and their abundance (y-axis). Each dot represents a unique spacer sequence; blue and red dots indicate a corresponding protospacer with an NGG or non-NGG PAM. Top and bottom plots indicate protospacers in the top and bottom strands of the ϕNM4γ4 DNA. The map as well as the different functions of the phage genes are indicated in between the plots. The raw data used to make this graph is in the Supplementary file. c, Weblogo showing the conservation of the 5’ flanking sequences of 10,000 protospacers randomly selected from the experiment shown in b. Absolute conservation of the NGG PAM was observed.