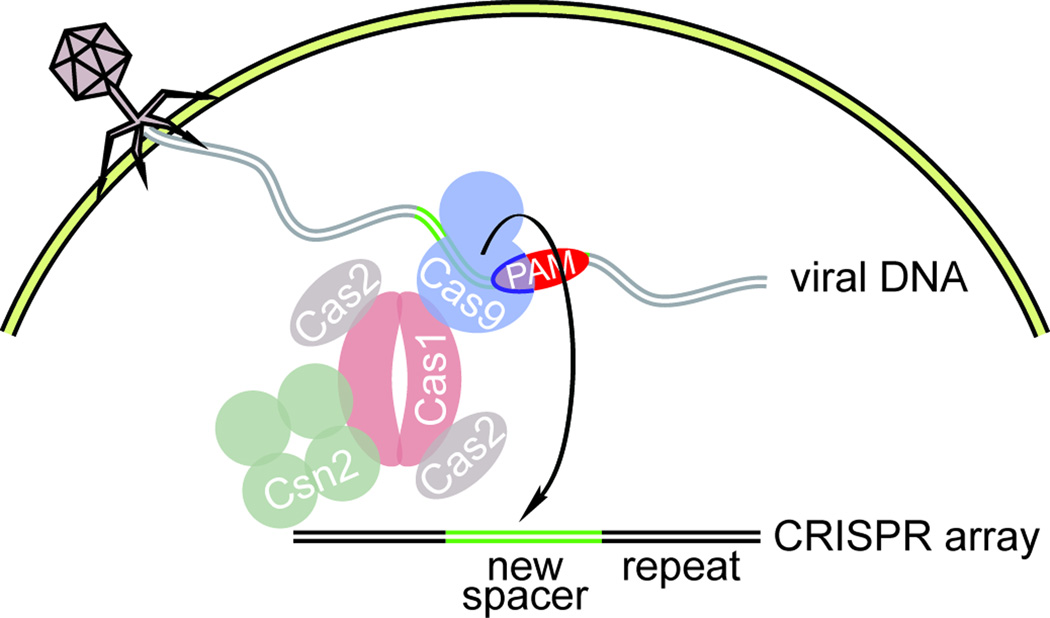
Extended Data Figure 6. A model for the selection of PAM-flanking spacers by Cas9.
After injection of the phage DNA an adaptation complex formed by Cas9, Cas1, Cas2 and Csn2 uses the Cas9 PAM binding domain to specify functional protospacers, i.e., that are followed by the correct PAM. It is not known how the protospacer sequence is extracted from the viral DNA to become a spacer. In the “cut and paste” model, a nuclease, possibly Cas1, cuts the viral DNA to generate the spacer. In the “copy and paste” model the protospacer sequence is copied first. Once loaded with the selected protospacer sequence, this complex promotes the integration of this sequence into the CRISPR array, thus becoming a new spacer. Previous studies demonstrated that Cas1 dimerizes and interacts with Cas2 (ref.13); Csn2 has been determined to form a tetramer34.