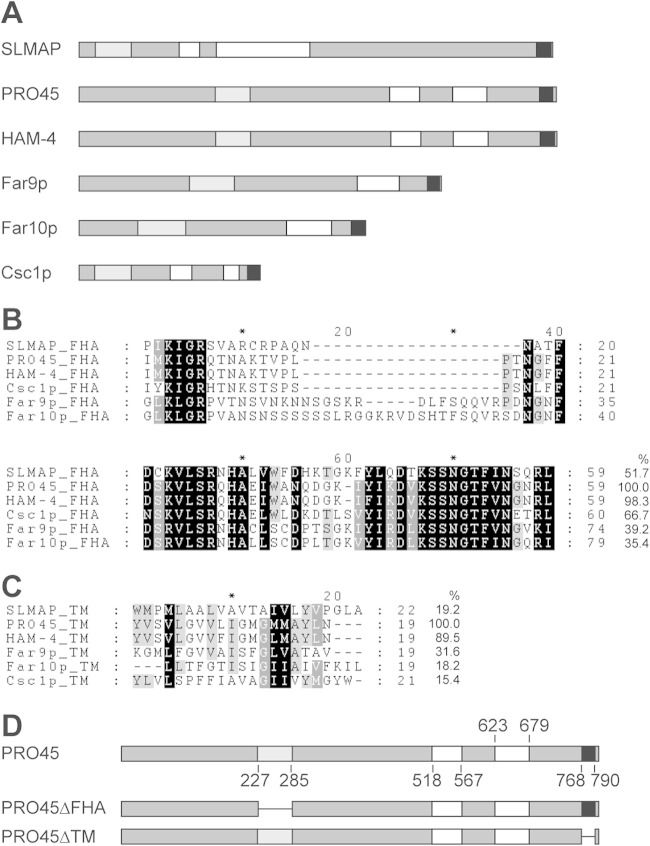
FIG 1.
SLMAP homologs. (A) Domain structure of SLMAP and fungal homologs. Proteins are depicted as medium gray bars. Light gray, white, and black bars, FHA, coiled-coil, and TM domains, respectively. Domains were analyzed by the use of the ELM (75) and TMPred (76) programs. (B, C) Amino acid alignment of FHA (B) and TM domains (C) from the indicated SLMAP homologs. Amino acid sequence identities (in percent) are given at the end of the alignments in relation to the S. macrospora PRO45 amino acid sequence. SLMAP, human SLMAP (GenBank accession number AAI14628.1); PRO45, S. macrospora PRO45 (GenBank accession number CCC07657.1); HAM-4, N. crassa HAM-4 (GenBank accession number EAA34844.2); Csc1p, S. pombe Csc1p (GenBank accession number NP_595762.1); Far9p, S. cerevisiae Far9p (GenBank accession number NP_010486.3), Far10p, S. cerevisiae Far10p (GenBank accession number EGA73760.1). (D) Domain structure of S. macrospora PRO45 derivatives PRO45ΔFHA and PRO45ΔTM. Numbers give the amino acid positions of the domains depicted in panel A.