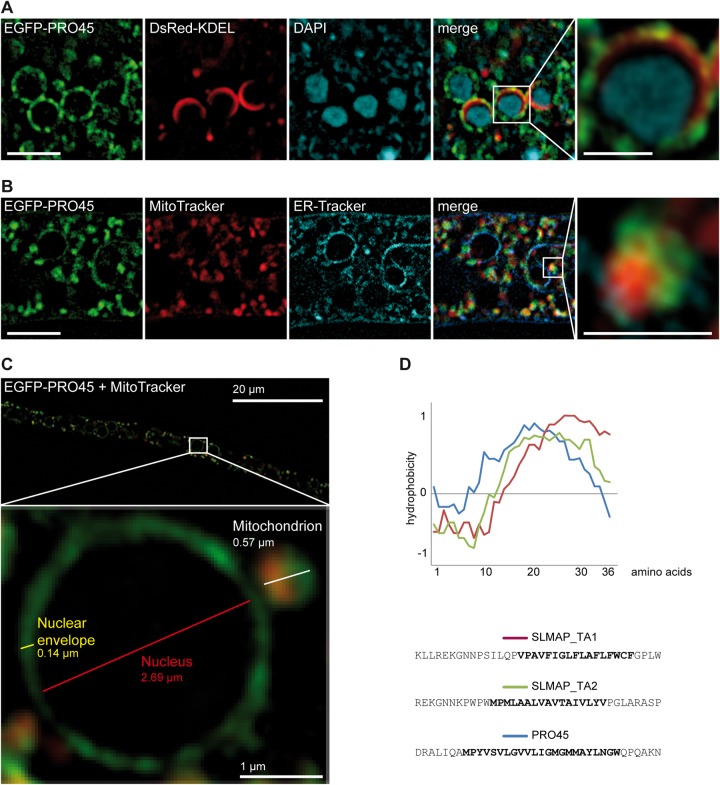
FIG 5.
Localization of PRO45 by SIM. (A) Strain N861(pEGFP-45, pDsREDKDEL) was stained with DAPI, indicating localization of PRO45 to the nuclear envelope. (B) Strain N506(pEGFP-45) was costained with MitoTracker and ER-Tracker, revealing the simultaneous association of PRO45 with the nuclear envelope and mitochondria. Bars, 5 μm (A and B) and 1 μm (insets). (C) Association of EGFP-PRO45 with the ER, the nuclear envelope, and mitochondria at high resolution in strain N506. Note the close association of the mitochondrion with the nuclear envelope. (D) Hydrophobicity of tail anchors (TA) of human SLMAP isoforms and S. macrospora PRO45. The graph represents the hydrophobicity of the last 36 amino acids of each protein, including the transmembrane domains (boldface sequences) and the very C-terminal amino acids. SLMAP_TA1 and SLMAP_TA2 represent two alternative splice isoforms of SLMAP showing different subcellular localizations to the ER (SLMAP_TA1) and to both the ER and mitochondria (SLMAP_TA2) (63). Hydrophobicity was calculated using the Eisenberg normalized scale with a window size of 9; the relative weight for window edges was 100.