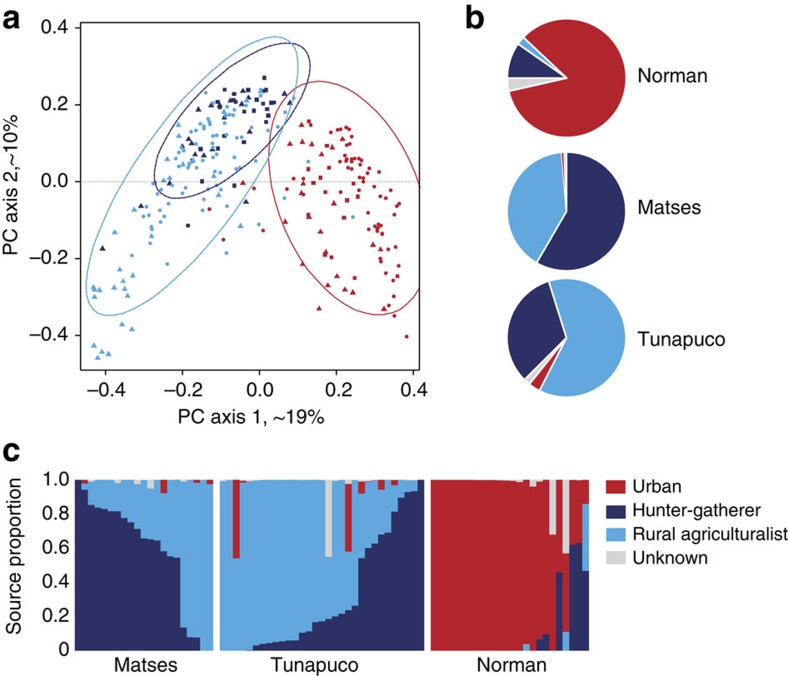
Figure 3. Comparison of the gut microbiomes of Matses, Tunapuco and Norman populations to published data from hunter-gatherer, rural agriculturalist and urban-industrial communities.
Analyses were performed on genus-level taxa tables rarefied to 4,000 reads per sample. (a) Principal coordinate analysis of Bray–Curtis distances generated from taxa tables summarized at the genus level. Proportion of variance explained by each principal coordinate axis is denoted in the corresponding axis label. Populations are colour coded by subsistence strategy. Data sets are represented by triangles (this study), circles (Yatsunenko et al.3), and squares (Schnorr et al.1). Ellipses correspond to 95% confidence boundaries for each of the three subsistence categories. (b) Results from Bayesian source-tracking analysis. Source contributions are averaged across samples within the population. (c) Results from Bayesian source tracking for individual samples.