Figure 4.
Characterization of TRABID Specificity
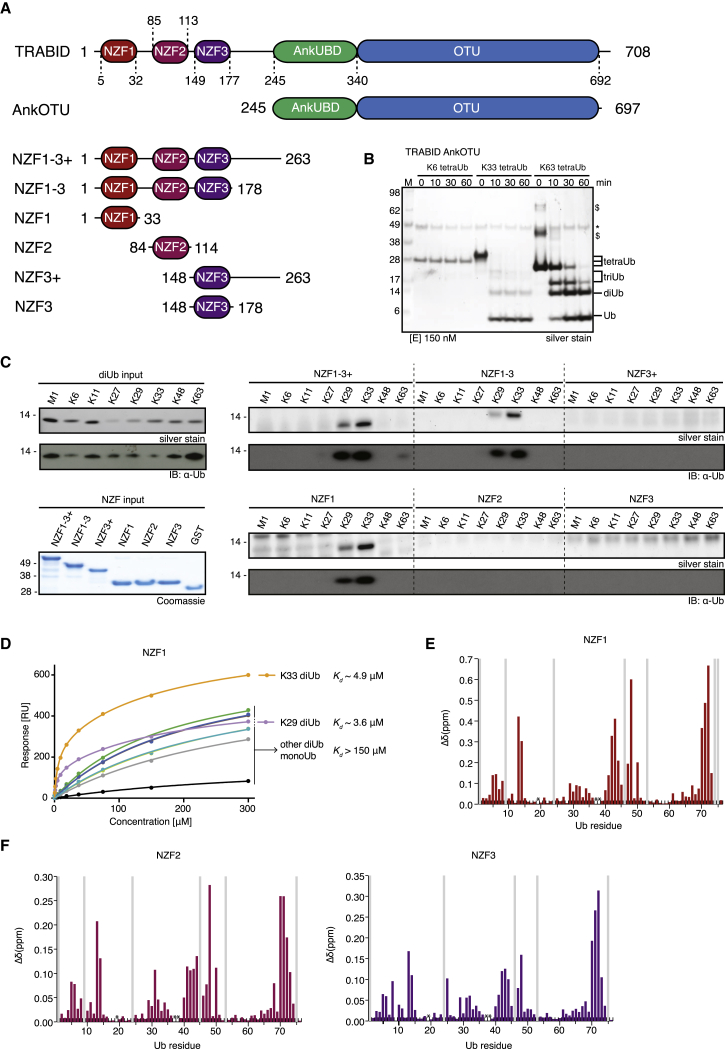
(A) Domain structure of human TRABID. The AnkOTU fragment has been characterized in detail in Licchesi et al. (2012). Boundaries of the NZF domain fragments analyzed here are shown.
(B) Deubiquitination assay of TRABID AnkOTU against K6-, K33-, and K63-linked tetraUb. See Figure S4A for a reaction at a lower DUB concentration.
(C) Pull-down analysis of NZF fragments with a panel of diUb covering all linkage types. Left: the input chains and GST-NZF constructs used. Right: pull-down analysis shown by silver stain and anti-Ub western blot. See Figure S4B for additional controls.
(D) SPR binding experiment of NZF1 to monoUb and the eight different diUb species with error bars representing SEs. Kd values derived from two experiments are shown. See Figure S4C for best-fit parameters and values of SEs.
(E) NMR analysis of isolated NZF1 binding to 15N-labeled monoUb. The chemical shift perturbation for Ub from binding to 600 μM of NZF1 is shown. Grey bars, exchange-broadened residues; asterisks, proline residues. See Figures S4E–S4G for titration data.
(F) NMR analyses as in (E) but for NZF2 and NZF3.