Figure 6.
Explaining the K29/K33 Specificity of TRABID NZF1
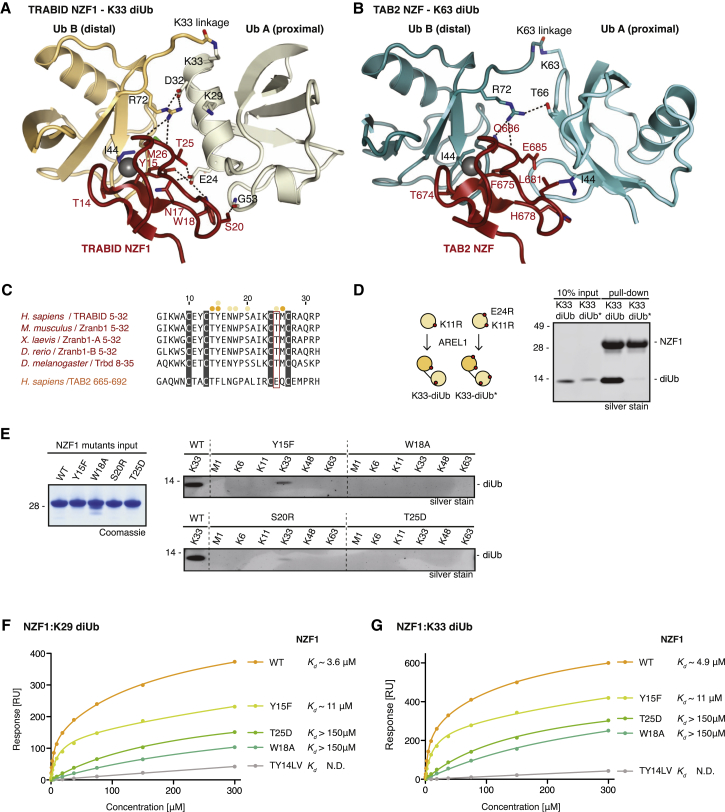
(A) Detailed view of the interactions between TRABID NZF1 (red) and K33-linked diUb (orange/beige). Interacting residues are labeled, and hydrogen bonds are shown as black dashed lines.
(B) As in (A) for the TAB2 NZF interaction with K63-linked diUb (cyan).
(C) Sequence alignment of TRABID NZF1 from a diverse range of species and human TAB2 NZF domains. Interacting residues are indicated with orange (distal Ub) and beige (proximal Ub) dots. Thr25 in TRABID NZF1 is replaced with Glu685 in TAB2 NZF, which would prevent K29/K33 binding in TAB2 NZF.
(D) Left: Ub chains were assembled into K33 diUb with AREL1 using K11R or K11R/E24R Ub. Right: pull-down assays with TRABID NZF1 and diUb variants.
(E) Pull-down assays as in Figure 4C for TRABID NZF1 mutants.
(F and G) SPR binding experiment of NZF1 and its mutants against K29 diUb (F) and K33 diUb (G) with the respective Kd values indicated. SEs from two experiments are shown as error bars. See Figure S6F for values of SEs and best-fit parameters.