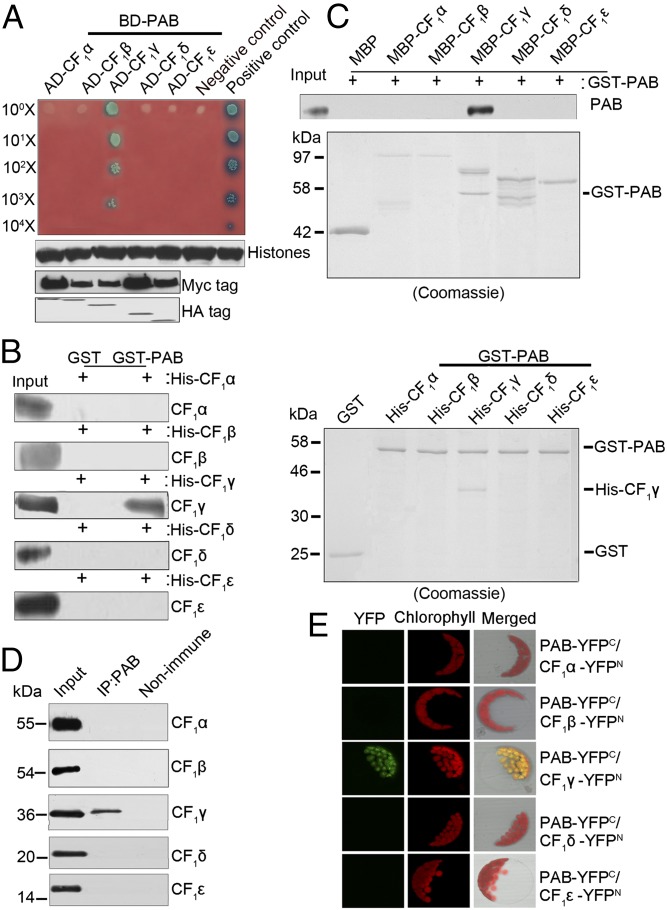
Fig. 2.
Interaction of PAB with CF1γ. (A) Yeast two-hybrid analysis of the interaction between PAB and CF1 subunits. The mature form of PAB was fused to the GAL4 DNA-binding domain (BD-PAB) as bait. CF1α, CF1β, CF1γ, CF1δ, and CF1ε were individually fused to the GAL4 activation domain (AD-CF1α, AD-CF1β, AD-CF1γ, AD-CF1δ, and AD-CF1ε) as prey. Yeast cells transformed with pGBKT7-53 and pGADT7-T were used as a positive control whereas those with pGBKT7-lam and pGADT7-T were a negative control. Immunoblot analysis of Myc and HA in yeast extracts was used to indicate the protein expression in the bait and prey plasmids, respectively. Immunoblot analysis of histone protein in yeast extracts was used as the estimation of yeast cell amounts. (B and C) Pull-down assays of the interaction between PAB and CF1 subunits. In B, PAB–glutathionine S-transferase (GST) fusion protein was constructed as bait. CF1α, CF1β, CF1γ, CF1δ, and CF1ε were individually fused to His as prey. In C, PAB-GST fusion protein was constructed as prey. CF1α, CF1β, CF1γ, CF1δ, and CF1ε were individually fused to maltose-binding protein (MBP) as bait. The bound proteins were eluted and analyzed by immunoblot analysis or Coomassie staining. (D) Coimmunoprecipitation (co-IP) analysis of the interaction between PAB and CF1 subunits. Arabidopsis total proteins were immunoprecipitated with nonimmune serum (right lane) or with antibodies against PAB (middle lane) and then analyzed by immunoblot with antibodies indicated on the right. Input (left lane) indicates that 100 μg total proteins was loaded on the gel. (E) Bimolecular fluorescence complementation analysis of the interaction between PAB and CF1 subunits. Plasmids encoding fusion constructs with the N- or C-terminal part of YFP (YFPN or YFPC, respectively) were transiently expressed in Arabidopsis protoplasts. YFP fluorescence indicates a direct interaction in planta localized to the chloroplasts.