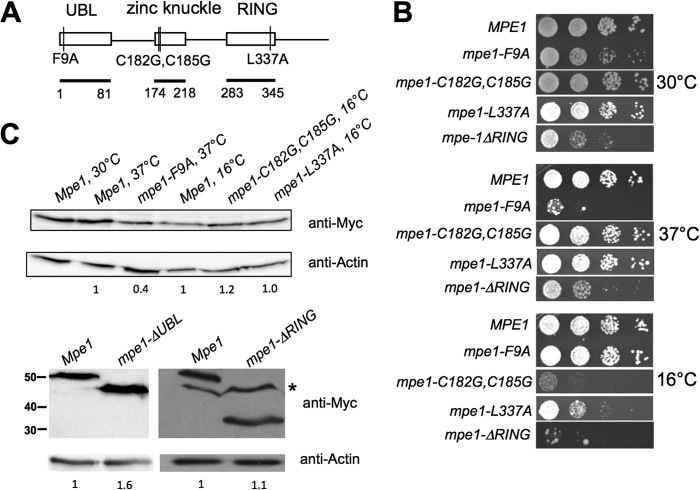
FIG 1.
MPE1 mutants show growth defects. (A) Schematic of mutations analyzed in this study, with conserved domains and their amino acid boundaries indicated. Amino acids 2 to 81 were deleted in the mpe1-ΔUBL construct, and amino acids 283 to 441 were deleted in the mpe1-ΔRING construct. UBL, ubiquitin-like domain. (B) Relative growth rates of mpe1 mutants at different temperatures. Strains were grown in liquid YPD, 10-fold serially diluted, spotted onto YPD plates, and incubated for 3 to 7 days at the indicated temperatures. (C) In vivo expression of the mpe1 mutant proteins. Cells containing a chromosomal deletion of the MPE1 gene but expressing Myc-tagged forms of wt MPE1 or mpe1 mutants on plasmids were first grown at 30°C and then, as indicated, shifted to 37°C for 2 h (mpe1-F9A) or to 16°C for 4 h (mpe1-C182G,C185G and mpe1-L337A). Extracts from these cells were analyzed by Western blotting using an antibody against the Myc epitope or against actin. For the nonviable mpe1-ΔUBL mutation, an extract was made from cells coexpressing an untagged wild-type Mpe1 protein, and only the truncated protein was detected by the anti-Myc antibody. The asterisk indicates a nonspecific band sometimes seen with the anti-Myc antibody. Numbers on the left of the bottom panel indicate the positions of molecular mass markers (in kilodaltons). The amount of mutant protein relative to the amount of wild-type Mpe1 was determined by using actin as a loading control and is indicated under each lane.