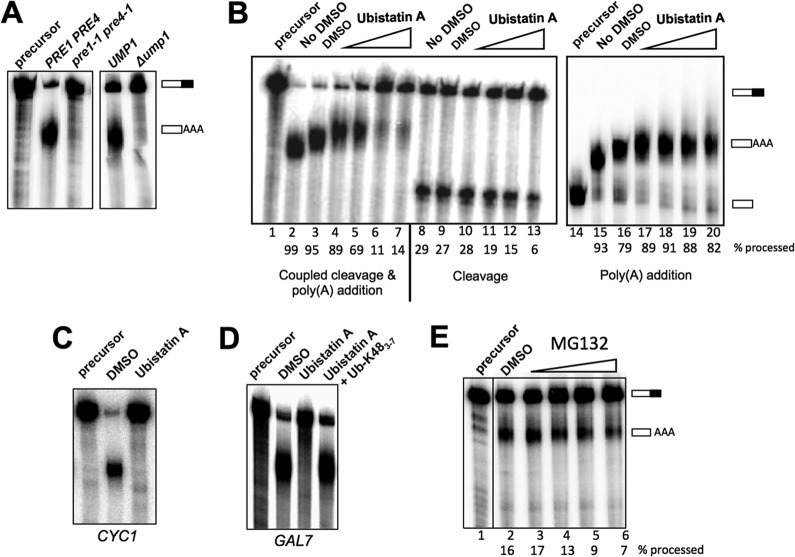
FIG 6.
Role of ubiquitination in mRNA 3′-end processing. (A) Processing extracts were prepared from wild-type, Δump1, and pre1-1 pre4-1 strains that had been grown at 30°C and then shifted to 37°C for 2 h. Processing reactions used GAL7-1 RNA, and RNAs were detected as described for Fig. 2. (B) Disruption of ubiquitin interactions inhibits 3′-end processing activity and, in particular, the cleavage step, in vitro. Whole-cell extracts were incubated with increasing amounts of ubistatin A in DMSO (5 μM, 20 μM, 100 μM, and 1 mM; indicated by the wedges above lanes 4 to 7, 10 to 12, and 17 to 20) or with (lanes 3, 9, and 16) or without (lanes 2, 8, and 15) DMSO for 10 min at room temperature before the addition of GAL7-1 RNA precursor to initiate the 3′-end processing assay, using the reaction conditions and the procedure for detection and quantitation of RNAs that are described in the legend to Fig. 2. (C) Ubistatin A inhibits the processing of the precursor containing the CYC1 poly(A) site. The coupled cleavage and poly(A) addition reaction was performed using ATP and radioactive precursor containing the CYC1 poly(A) site. (D) Ubistatin A inhibition is rescued by polyubiquitin. Seventy micromolar ubistatin A was preincubated with 100 μM K48-linked tetraubiquitin for 10 min at room temperature and then incubated with the whole-cell extract for an additional 10 min before the addition of the GAL7-1 RNA. (E) MG132 inhibits 3′-end processing activity. The processing extract was incubated with MG132 in DMSO (1 μM, 10 μM, 100 μM, and 1 mM) or with DMSO alone for 10 min at room temperature before the addition of GAL7-1 RNA precursor to initiate the 3′-end processing assay, using the reaction conditions and the procedure for detection and quantitation of RNAs that are described in the legend to Fig. 2. The samples were loaded onto the same gel, and the intervening lane was removed as demarked by the line between lanes 1 and 2.