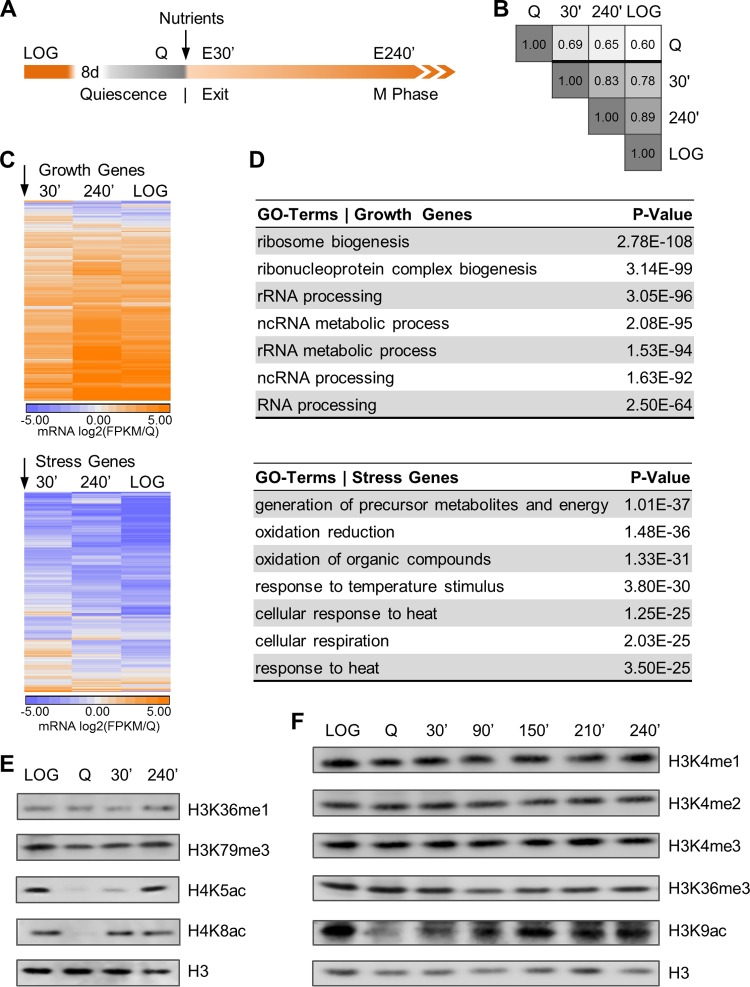
FIG 1.
Nutrient availability stimulates histone acetylation and dramatic change in gene expression but does not alter global histone methylation levels. (A) Schematic of yeast culturing and quiescence exit protocol. Cells are grown in rich medium (LOG) until carbon exhaustion and are then transferred to water to ensure a completely starved, homogeneous quiescent cell population (Q) prior to nutrient-induced quiescence exit on day 8. The arrow indicates nutrient feeding and subsequent sampling 30 min and 240 min after nutrient feeding (E30′ and E240′, respectively). (B) Pearson correlation matrix of mRNA transcriptomes. In nutrient-induced cell cycle reentry, changes in global gene expression advance genome-wide transcription to the growth profile of cycling cells. (C) mRNA sequencing data show that nutrient feeding (arrows) induces vast transcriptional reprogramming in the transition from quiescence to cellular growth. Following nutrient replenishment, 475 growth genes are drastically upregulated (orange), whereas a core set of 463 stress genes are rapidly downregulated (blue). Data are clustered by time points into columns and rows; rows represent genes, which are ranked by FPKM (where FPKM stands for fragments per kilobase of transcript per million mapped reads) values normalized to quiescence sample FPKM and log2 transformed. (D) GO term enrichment analysis shows that growth genes are involved in processes critical to growth. Stress genes cluster in classic stress response categories. (E and F) Western blot analysis of lysates from yeast over the course of quiescence exit reveals that total levels of histone acetylation are diminished in quiescent cells and rapidly increase following nutrient replenishment. In contrast, global histone methylation levels remain stable throughout quiescence establishment and do not increase during quiescence exit.