FIG 1.
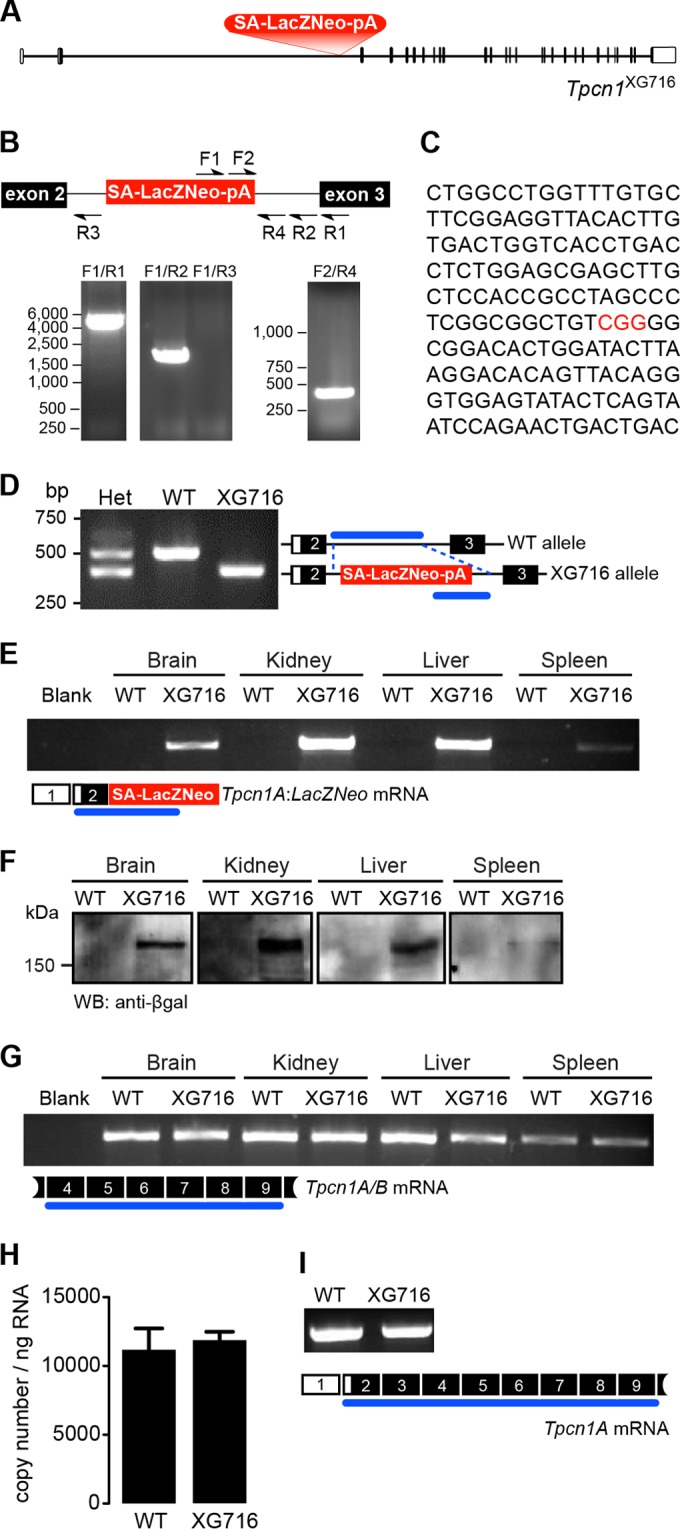
The gene trap mutation in Tpcn1XG716 does not abolish Tpcn1 expression. (A) Schematic representation of Tpcn1XG716 gene structure based on the gene with GenBank accession number NM_145853.2. Red block, gene trap sequences (SA, splice acceptor; LacZNeo, chimeric β-galactosidase and neomycin phosphotransferase II sequences; pA, polyadenylation signal); vertical segments, exons; unfilled sections, UTRs. (B) PCR strategy for determination of the insertion site of gene trap vector pGT1Lxf in the Tpcn1XG716 allele. Several combinations of forward (F) and reverse (R) primers were used to narrow the location of the gene trap vector within the intron. (C) The product from PCR with primers F2/R4 was cloned into pCRIITOPO (Invitrogen) and sequenced, and the insertion site of the gene trap within the intron was determined to be between 2,754 and 2,757 bp upstream of exon 3 (marked in red). (D) Genotyping for WT and Tpcn1XG716 alleles. Numbered white and black blocks, noncoding and coding exons, respectively; blue lines, PCR-amplified regions; Het, heterozygote. (E) RT-PCR analysis of the expression of the chimeric trapped Tpcn1XG716 transcript (Tpcn1A-LacZ-Neo) from WT and homozygote Tpcn1XG716 animal tissues. Blank, a reaction with no RNA. (F) Immunoblotting analysis of chimeric TPC1A–β-galactosidase–neomycin phosphotransferase II protein in tissue homogenates from WT and homozygote Tpcn1XG716 animals using an anti-β-galactosidase (anti-β-gal) antibody. WB, Western blotting. (G) Expression of Tpcn1A/B determined by probing for transcript regions downstream from the gene trap insertion in tissues from WT and homozygote Tpcn1XG716 animals. (H) Copy number of Tpcn1A/B transcripts in liver from WT and homozygote Tpcn1XG716 animals determined by RT-qPCR. Bars correspond to the mean ± SEM for 4 to 8 animals. (I) Expression of the Tpcn1A transcript in the liver resulting from skipping of the gene trap cassette.
