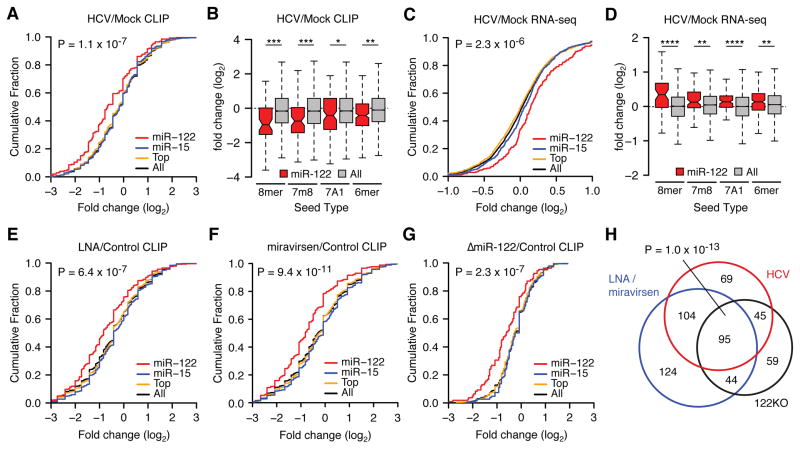
Figure 3.
HCV infection de-represses endogenous miR-122 targets. (A) Cumulative density function (CDF) of the log2 fold change in CLIP binding between infected and uninfected cells for all 3′UTR clusters containing indicated 7–8mer seeds by family, from triplicate experiments. “Top” refers to the top 10 miRNA families, exclusive of miR-122. “All” refers to the top 50 miRNA families, inclusive of miR-122. Two-sided K-S test p-value between miR-122 and all targets shown. (B) The mean log2 fold change (± ranges) in CLIP binding on miR-122 3′UTR targets versus all targets during HCV infection broken down by seed type. (C) A CDF plot during HCV infection as in (A) but measuring target mRNA expression via RNA-Seq, from duplicate experiments at 72hrs post-infection. Targets with more than one miRNA binding site were collapsed such that no gene is represented more than once per category. (D) The mean log2 fold change (± ranges) in mRNA expression of CLIP targets during HCV infection broken down by seed type. (E–G) CDF plot as in (A), between treatment over control cells with LNA122 (E) or miravirsen (F) at 30nM or genetic deletion (G) of miR-122 (ΔmiR-122), each from triplicate experiments. (H) Proportional Venn diagram showing the overlap of miR-122 targets with reduced CLIP binding across ΔmiR-122, LNA or miravirsen treatment, and HCV infection conditions. Hypergeometic p-value of overlap shown. Asterisks: ****P<0.0001, ***P<0.001, **P<0.01, *P<0.05, two-sided Mann-Whitney U-test. See also Figures S2 and S4.