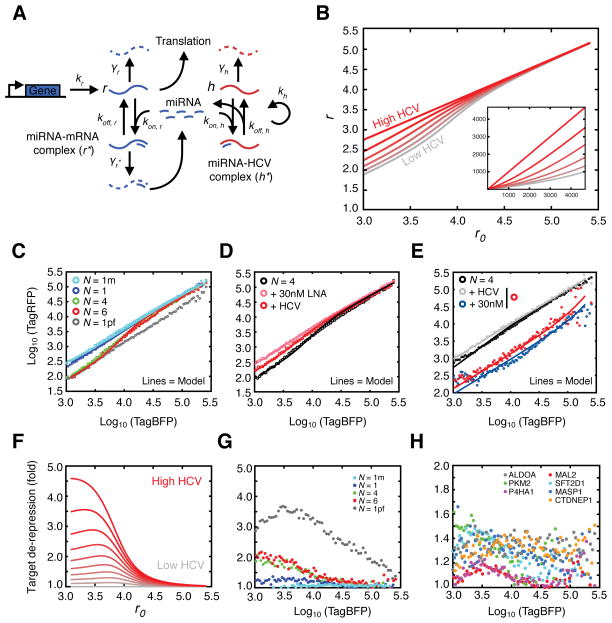
Figure 6.
Quantitative modeling of miR-122 sequestration by HCV. (A) Illustration of model reactions for miR-122 dynamics, including transcription and translation of a target mRNA, binding to miR-122 and decay of mRNA species. HCV RNA can replicate, be degraded, or bind miR-122, functionally sequestering miR-122 and leading to de-repression of mRNA targets. (B) Increasing amounts of HCV (or a relative increase in binding strength at miR-122 sites) leads to changes in single-cell gene expression as compared to unregulated targets, with stronger effects at the low mRNA expression levels. Parameters used are fitted from data in (C). Each curve, from top to bottom, represents a reduction in the miRNA pool by 20%. Inset displays model on a linear scale. (C) Model fitting of the steady state approximation to experimental data while increasing the number of binding sites corresponding to changes in total binding strength. (D) Model fitting for the N=4 case showing a 50% reduction in the miRNA pool by HCV modeled by a proportional change in the theta parameter. (E) Model fitting for the N=4 construct under 30nM miR-122 mimic addition ± HCV infection. (F) Increasing HCV:miR-122 binding strength or HCV RNA abundance results in functional de-repression of miR-122 targets. The curves (top to bottom) represent 10 percentage-points increases in the available miR-122 pool (10% to 100% availability). (G–H) Experimental HCV induced derepression of synthetic miR-122 binding site constructs (G) or endogenous 3′ UTRs with miR-122 binding sites (H). See also Figure S5 and S6.