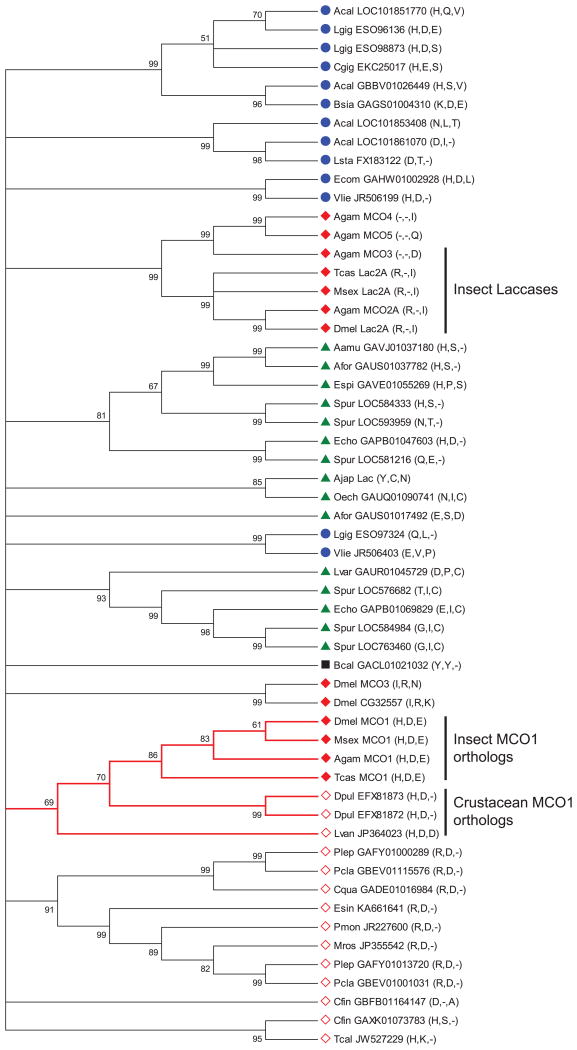
Figure 7.
Phylogenetic analysis of MCO genes from insects and other invertebrates. A bootstrap consensus tree constructed by the neighbor-joining method is shown. The numbers above the branches are bootstrap values expressed as a percentage. All branches with bootstrap values less than 50 were collapsed. A cluster of putative MCO1 orthologs is indicated by red branches. The residues corresponding to His374, Asp380 and Glu552 in DmMCO1 are listed after each gene name. Symbols indicate sequences from crustaceans (open red diamond), echinoderms (green triangle), insects (closed red diamond), molluscs (blue circle), or rotifers (black square). Abbreviations used are: Acal, Aplysia californica; Ajap, Apostichopus japonicas; Aamu, Asterias amurensis; Afor, Asterias forbesi; Bsia, Bithynia siamensis goniomphalos; Bcal, Brachionus calyciflorus; Cfin, Calanus finmarchicus; Cqua, Cherax quadricarinatus; Cgig, Crassostrea gigas; Dpul, Daphnia pulex; Espi, Echinaster spinulosus; Ecom, Elliptio complanata; Esin, Eriocheir sinensis; Echo, Evechinus chloroticus; Lvan, Litopenaeus vannamei; Lgig, Lottia gigantean; Lsta, Lymnaea stagnalis; Lvar, Lytechinus variegatus; Mros, Macrobrachium rosenbergii; Oech, Ophiocoma echinata; Pmon, Penaeus monodon; Plep, Pontastacus leptodactylus; Pcla, Procambarus clarkii; Spur, Strongylocentrotus purpuratus; Tcal, Tigriopus californicus; Vlie, Villosa lienosa.