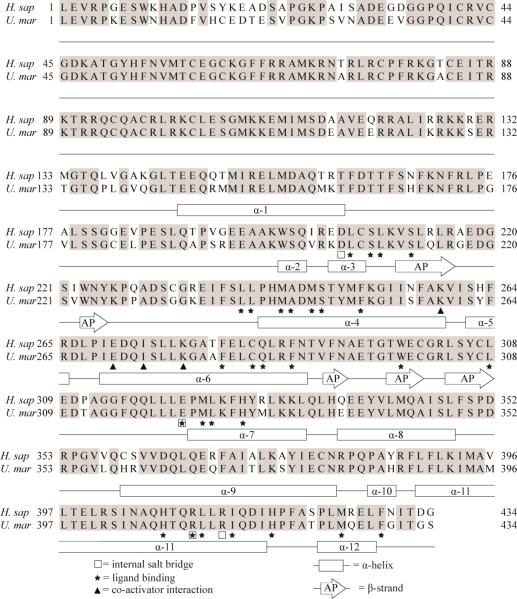
Figure 3. Alignment of hPXR and pbPXR supplemented with known secondary structure and ligand-binding residues.
Human PXR structures (Chrencik et al. 2005; Teotico et al. 2008; Watkins et al. 2001; Watkins et al. 2003; Xue et al. 2007a; Xue et al. 2007b) was used to supplement a hPRX and pbPXR amino acid sequence comparison with information of residues known to be involved in ligand binding, secondary structures, salt bridge and SRC-1 interaction. Rectangular boxes indicate α-helices (α) and arrows anti β-strands that form an antiparallel (AP) sheet. Stars indicate ligand-binding residues, squares residues involved in salt-bridges and triangles residues interacting with co-activator (SRC-1).