FIG 2.
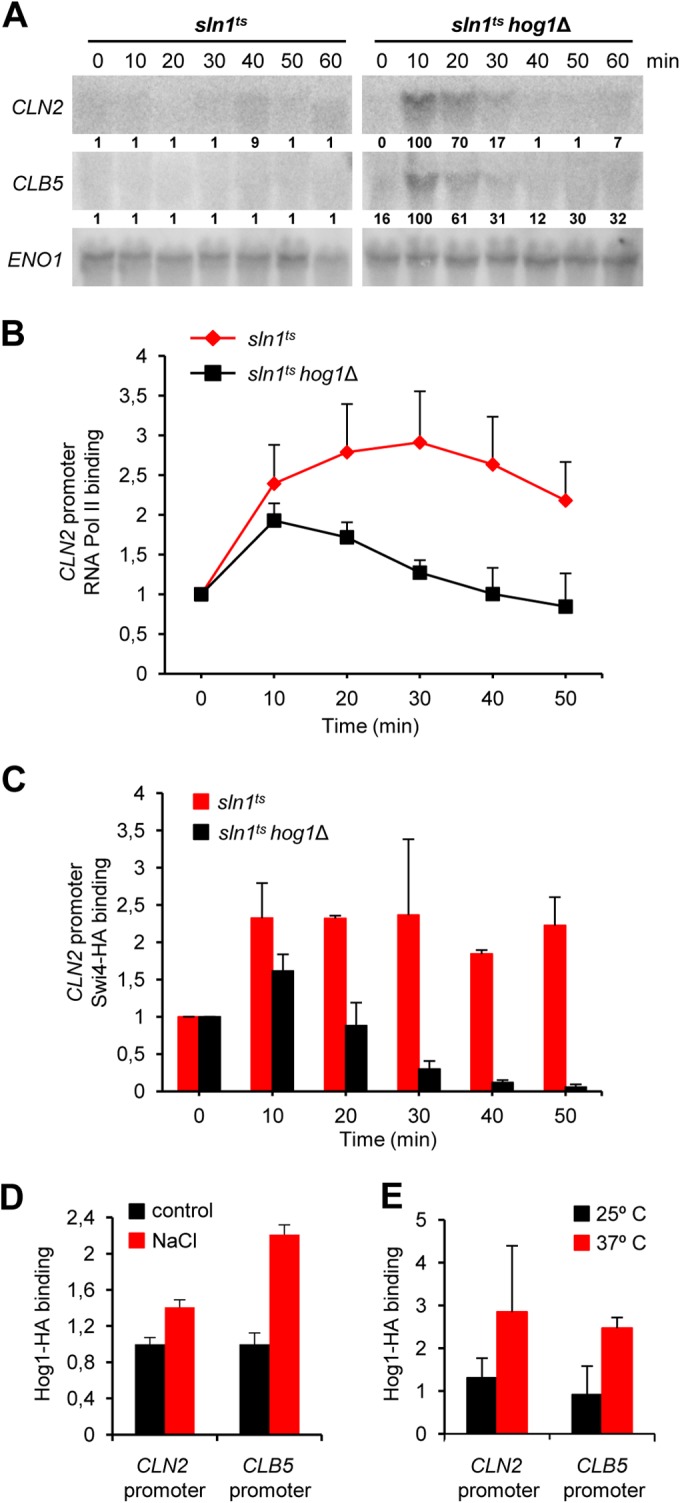
Hog1 is involved in transcriptional downregulation of G1 cyclins. (A) Constitutive activation of the HOG pathway downregulates G1-specific cyclin transcription. sln1ts or sln1ts hog1Δ mutant cells were synchronized in G1 with pheromone and then released into YPD medium at 37°C. RNA was extracted from samples at the times indicated and analyzed by Northern blotting with CLN2, CLB5, and ENO1 probes. Quantification relative to the loading control of each mRNA is depicted below each lane. A value of 100% is assigned to the maximum level of a given transcript. (B) RNA Pol II downregulation depends on Hog1. sln1ts and sln1ts hog1Δ mutant cells were synchronized with α-factor and released into YPD medium at 37°C. RNA Pol II was immunoprecipitated with anti-Rpb1 antibodies at the times indicated after release, and associated chromatin-DNA was analyzed by real-time PCR with primers specific for the CLN2 promoter region. Graphs represent the averages ± the standard deviations from three independent experiments. (C) Swi4 is retained at the CLN2 promoter upon Hog1 activation. Swi4 was tagged with HA in an sln1ts or sln1ts hog1Δ background. The resulting strains were synchronized with α-factor and released into YPD medium at 37°C. Swi4-HA was immunoprecipitated with anti-HA antibodies at the time points indicated after release. Associated chromatin-DNA was analyzed by real-time PCR with primers specific for the CLN2 promoter. Bars represent the averages ± the standard deviations from three independent experiments. (D, E) Hog1 binds to the promoter regions of CLN2 and CLB5. Wild-type cells were incubated for 10 min in the presence or absence (control) of 0.4 M NaCl (D), or sln1ts mutant cells were incubated at a nonpermissive temperature (37°C) for 30 min (E). HA-tagged Hog1 was used for chromatin immunoprecipitation in ChIP assays to monitor its association with the CLN2 and CLB5 promoters as described previously (49). Bars represent the averages ± the standard deviations from three independent experiments.
