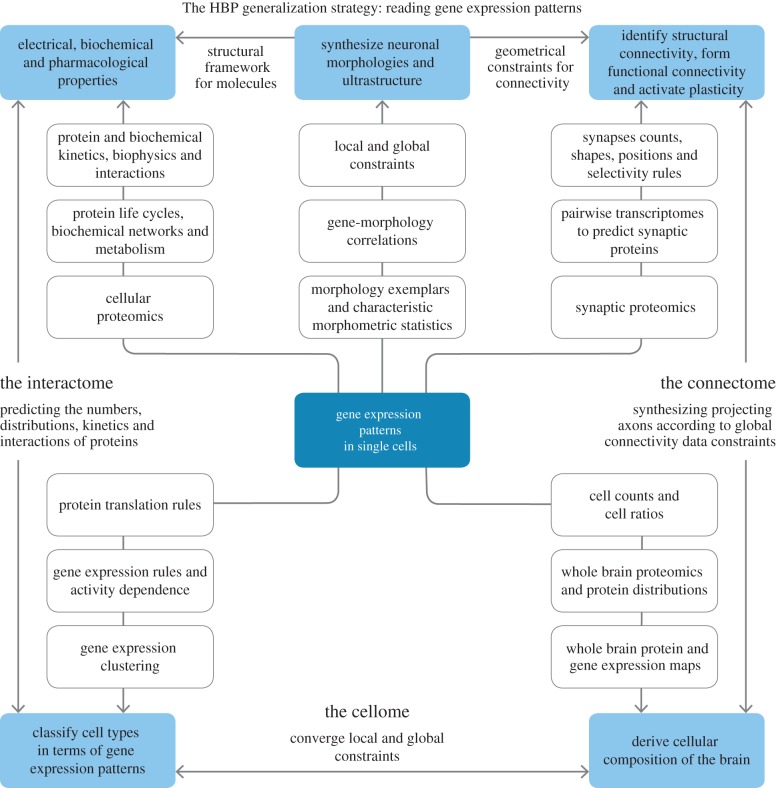
Figure 3.
A generic, futuristic schema for predictive reconstruction of a particular instance of a brain (a brain belonging to a specific species with a specific age and gender, and a specific genome). The fundamental data required for future brain mapping come from analysis of gene expression patterns in single cells. These data make it possible to consider clusters of cells with similar patterns as genetic cell types. From this starting point, many predictions become possible. By correlating genetic cell types with morphological properties and with constraints on cellular local and global locations (position), it becomes possible to synthesize three-dimensional models of all cell morphologies (including morphologies that have not yet been recorded) and to validate these models against known morphologies. The same data are used to predict cellular composition (at a genetic/molecular level). This is derived by finding the distribution of genetic types that best fits existing whole brain single gene expression maps (in terms of non-negative least squares). The predicted distribution is then validated against whole brain maps of cellular protein expression, thus yielding a complete cellome (the set of all cell types and their respective numbers and distributions). The connectome is derived using brain regional and inter-regional projection data, combined with known principles of synaptic connectivity at the micro-, meso- and macro-levels validated against electron microscopic image stacks from each brain region. Correlations in single-cell transcriptomes of pairs of neurons of known genetic type, and knowledge of the way mRNA is translated into proteins is used to predict the anatomical, physiological, molecular and plasticity properties of the synapses they form and is validated against whole brain maps of synaptic proteins, known synaptic physiology and plasticity. Single-cell gene expression patterns, protein translation principles and protein addressing principles are used to populate and distribute proteins (ion channels, receptors, etc.), peptides, metabolites and other biomolecules within neurons, glia and synapses. Predictions are validated against cellular level gene and protein staining patterns. Reaction kinetics are predicted from fundamental structure–function principles determined in dynamical simulations of molecular interactions. The results can be used to predict a complete interactome and validated against known interactions. At each stage of a reconstruction simulation, results are validated against biophysical, physiological and pharmacological data and where appropriate, against data from cognitive and behavioural studies.