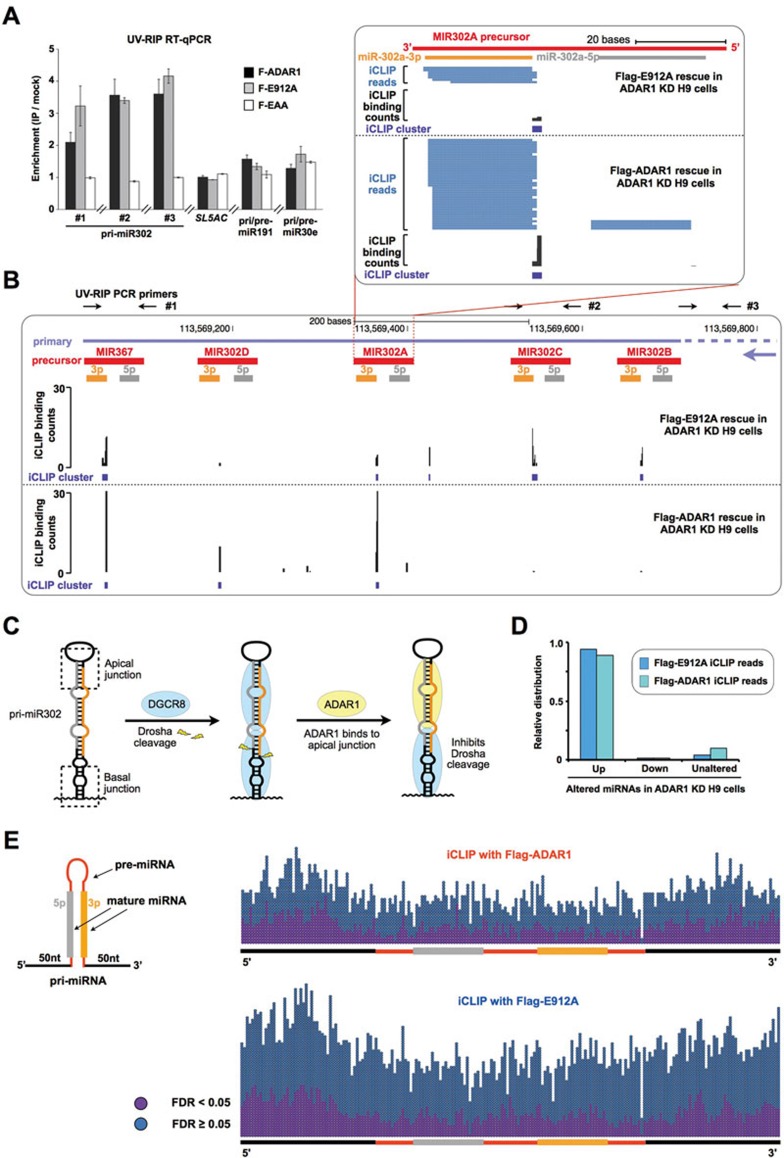
Figure 6.
ADAR1 binds to pri-miR302s and interferes with processing. (A) UV crosslinking RIP confirms that both Flag-ADAR1 and Flag-E912A, but not Flag-EAA, are associated with the polycistronic primary transcript of miR302s. Note that non-altered miRNAs, such as miR191 and miR30e, were not precipitated by ADAR1 proteins. qPCR primers for pri-miR302 detection are labeled as black arrows in B. Error bars represent ± SD of triplicate experiments. (B) iCLIP-seq defines that Flag-ADAR1- and Flag-E912A-binding sites are located within pri-miR302 hairpins. Individual Flag-ADAR1 and Flag-E912A iCLIP-seq reads (in blue) or iCLIP binding counts (in black) were aligned to pri-miR302s, with the mature miRNA boundaries depicted below (orange and gray bars). The iCLIP clusters are depicted as purple rectangles. (C) Two DGCR8 dimers clamp a pri-miRNA hairpin and activate Drosha cleavage (middle)52. The binding of ADAR1 to hairpins of pri-miR302s competes with DGCR8 binding, resulting in an inhibitory effect on pri-miR302 processing (right). (D) The majority of Flag-ADAR1 and Flag-E912A iCLIP reads are located in pri-miRNA regions of the upregulated miRNA targets after ADAR1 knockdown in H9 cells. (E) Flag-ADAR1 and Flag-E912A bind to pri-miRNAs with no sequence specificity. iCLIP clusters, significant (purple points) or non-significant (blue points), were scattered evenly on pri-miRNAs for both Flag-ADAR1 (right panel, top) and Flag-E912A (right panel, bottom). Also see Supplementary information, Figures S5, S6 and Table S6.