Figure 1.
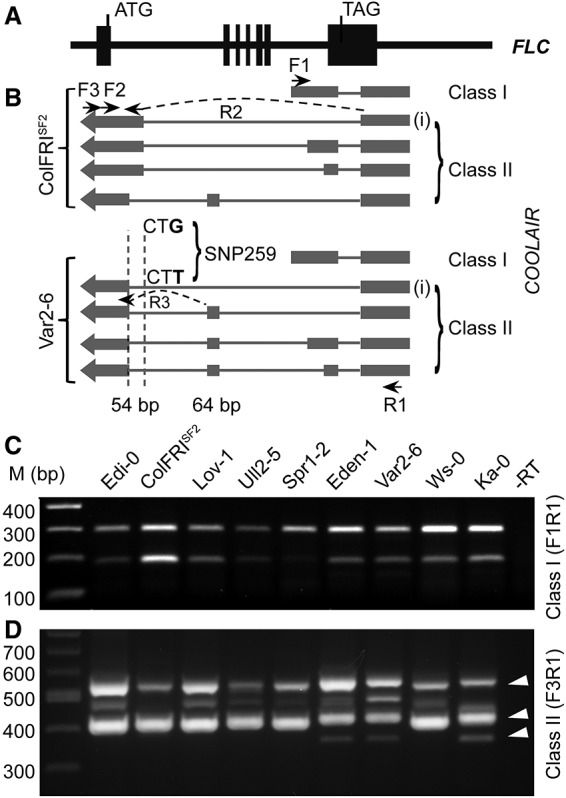
SNP259 in FLC influences COOLAIR splicing. (A,B) Schematic illustration of alternative COOLAIR splicing and polyadenylation in Col FRISF2 (FRI from SF-2 accession) and Var2-6. (Black boxes) FLC exons; (grey boxes) COOLAIR exons. The position of SNP259 (Col-0/Var2-6: G/T) is indicated; the nucleotides CT show the splice acceptor site for COOLAIR class II transcripts (“i” is marked). Arrows show PCR primers used to assay splicing; arrows plus a dashed line show primers that cover an exon–intron junction. The 54-base-pair (bp) shift of the COOLAIR acceptor site is highlighted with two vertical dashed lines, and the 64-bp additional exon is marked. (C) The COOLAIR class I splicing pattern is the same in a set of accessions representing the different FLC haplotypes. The RT–PCR primers F1 and R1 are as shown in B. For class I, the lower band is due to mispriming of the F1 oligo and represents unspliced class I COOLAIR. (D) COOLAIR class II is alternatively spliced in a set of accessions representing different FLC haplotypes. The RT–PCR primers F3 and R1 are as shown in B. The arrowheads indicate the characteristic COOLAIR splicing changes in accessions containing the SNP259T polymorphism.
