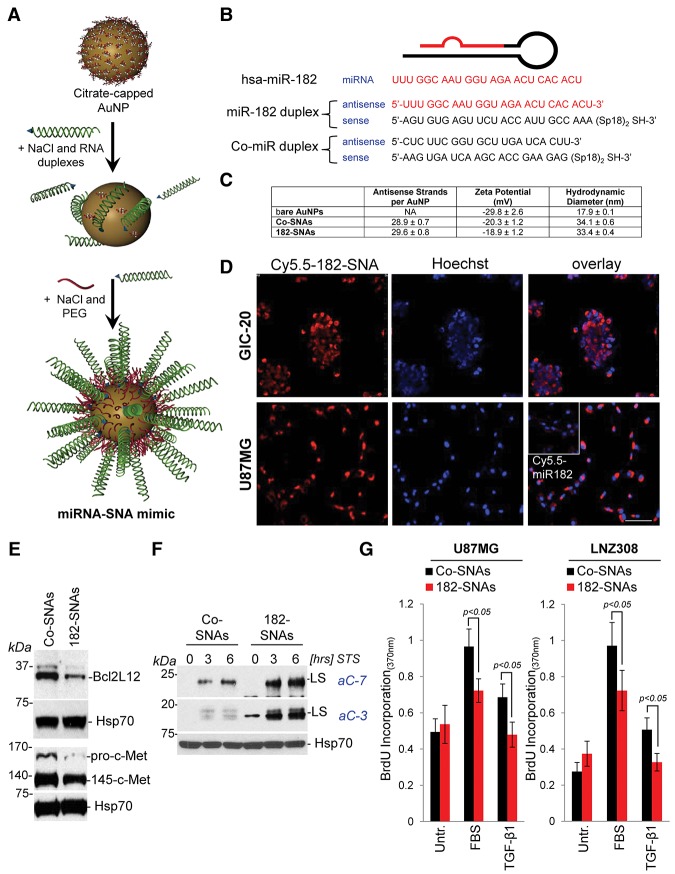
Figure 5.
miR-182-based SNAs penetrate glioma cells, robustly down-regulate miR-182 target genes, and phenocopy cellular effects of lipoplex-delivered miR-182 sequences. (A) miR-182 or Co-miR–RNA duplexes were hybridized to citrate stabilized gold nanoparticles (AuNPs) via thiol-gold bond and passivated with polyethylene glycol-Thiol (mPEG-SH). (B) Sequence of miR-182 and Co-miR duplexes. (C) Physico–chemical characterization of SNAs as outlined in the Materials and Methods. (D) Confocal images of U87MG and GIC-20 treated with Cy5.5-labeled SNAs or free miR-182 sequences (inset) and counterstained with Hoechst dye to visualize the nuclei. Bar, 50 μm. (E) U87MG cells were treated with 10 nM Co-SNA or 182-SNAs for 48 h, and protein levels of Bcl2L12 and c-Met were assessed by Western blotting. (F) Western blot analysis for active caspase-3 and caspase-7 in U87MG cells that were transfected with 10 nM Co-SNAs or 182-SNAs for 48 h and subsequently treated with 0.5 μM STS for the indicated periods of time. (G) BrdU incorporation assays in the presence of 10% FBS or 10 ng/mL TGF-β1 in U87MG and LNZ308 treated with 10 nM Co-SNAs or 10 nM 182-SNAs for 24 h.