Figure 1.
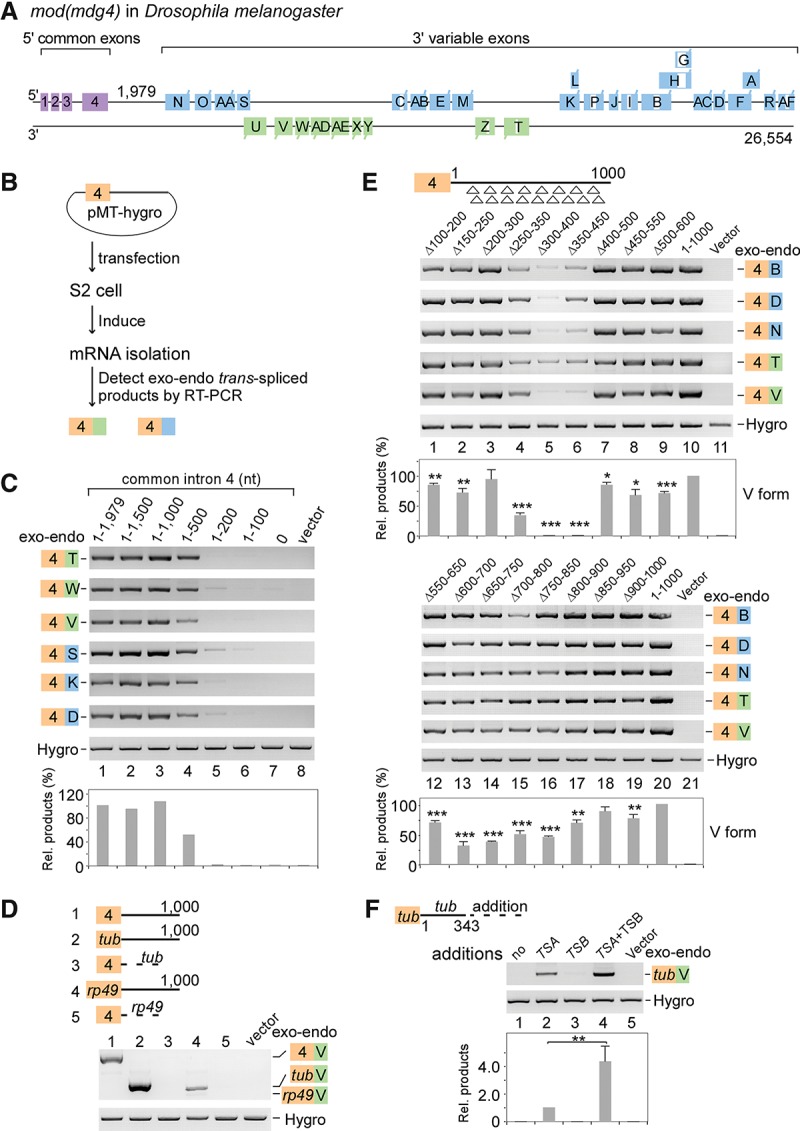
Two intronic RNA sequences are critical for trans-splicing of mod(mdg4). (A) Schematic of the mod(mdg4) gene locus in Drosophila melanogaster. Exons and introns from 3′ transcripts are designated according to FlyBase, and the sequence between exon 4 and the first 3′ exon N is defined as the last 5′ intron (intron 4). (B) Schematic of a trans-splicing system in Drosophila S2 cells. (C) Truncation assays of intron 4 reveal that two regions are important for trans-splicing activity. Relative trans-spliced products are averages from six isoforms that were normalized to loading controls and the full-length intron 4. (D) Replacing intron 4 of mod(mdg4) with other introns abolishes trans-splicing activity. Sequences of other genes include exon 1–intron 1 (without the 3′SS) from tubulin (tub) and rp49. (E) Trans-splicing activity of tiled 101-nt deletions in intron 4. Relative trans-spliced products of mod(mdg4) V forms were quantitated and normalized to the intact intron 4 (mean ± SEM; n = 3). For shorter tiled deletions, see Supplemental Figure S1C. (F) TSA RNA is sufficient to promote trans-splicing, while TSB RNA enhances the activity. TSA and TSB RNAs are fragments of the 330–500 nt and 650–800 nt in mod(mdg4) intron 4, respectively. Relative enhancement by TSB was normalized to TSA alone (mean ± SEM; n = 3). (Purple boxes) Chromosomal 5′ exons; (blue boxes) 3′ exons from the same DNA strand; (green boxes) 3′ exons from the opposite strand; (brown boxes) exon 4 on plasmids; (white boxes) internal cis-spliced intron in 3′ exons; (Hygro) hygromycin B. Boxes and labels are used similarly in other places unless otherwise indicated. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001.
